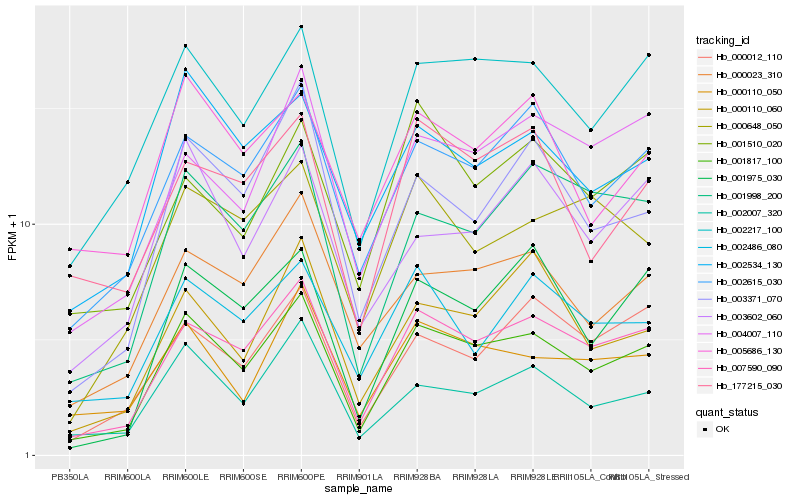
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_100 |
0.0 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 2 |
Hb_007590_090 |
0.0604516224 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 3 |
Hb_002217_100 |
0.0873966543 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_002615_030 |
0.0979104804 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 5 |
Hb_000023_310 |
0.1064512622 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 6 |
Hb_002007_320 |
0.1076649522 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 7 |
Hb_001998_200 |
0.1078326051 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 8 |
Hb_002534_130 |
0.1132189829 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 9 |
Hb_001975_030 |
0.1152361465 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 10 |
Hb_000110_050 |
0.1166837 |
- |
- |
- |
| 11 |
Hb_003371_070 |
0.1179283926 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000110_060 |
0.120015228 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 13 |
Hb_000012_110 |
0.1222274272 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 14 |
Hb_004007_110 |
0.1223332297 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 15 |
Hb_002486_080 |
0.129319466 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 16 |
Hb_003602_060 |
0.1294065262 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 17 |
Hb_177215_030 |
0.1314930212 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_005686_130 |
0.1323879378 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 19 |
Hb_000648_050 |
0.1346475584 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001510_020 |
0.1379659179 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |