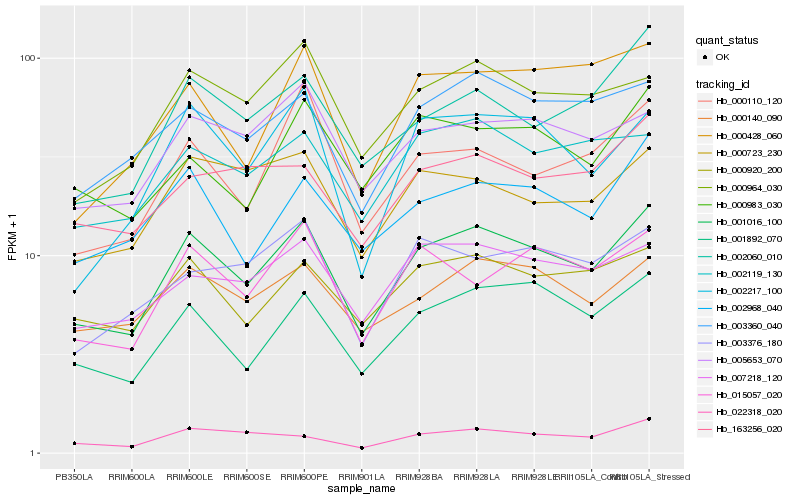
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001016_100 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 2 |
Hb_001892_070 |
0.0824273848 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 3 |
Hb_000920_200 |
0.0916712578 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003360_040 |
0.0923916242 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 5 |
Hb_000140_090 |
0.0939403154 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 6 |
Hb_002968_040 |
0.0947013713 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 7 |
Hb_000964_030 |
0.0949567493 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 8 |
Hb_005653_070 |
0.0960552001 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 9 |
Hb_007218_120 |
0.0964041066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 10 |
Hb_015057_020 |
0.0977373648 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 11 |
Hb_002119_130 |
0.0992577409 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 12 |
Hb_000723_230 |
0.1016452602 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 13 |
Hb_002060_010 |
0.1023916575 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002217_100 |
0.1026841092 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000983_030 |
0.1053322864 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 16 |
Hb_022318_020 |
0.1061257064 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 17 |
Hb_000428_060 |
0.1077668241 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_163256_020 |
0.1089005281 |
- |
- |
fructokinase [Manihot esculenta] |
| 19 |
Hb_003376_180 |
0.1089200002 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 20 |
Hb_000110_120 |
0.1091818069 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |