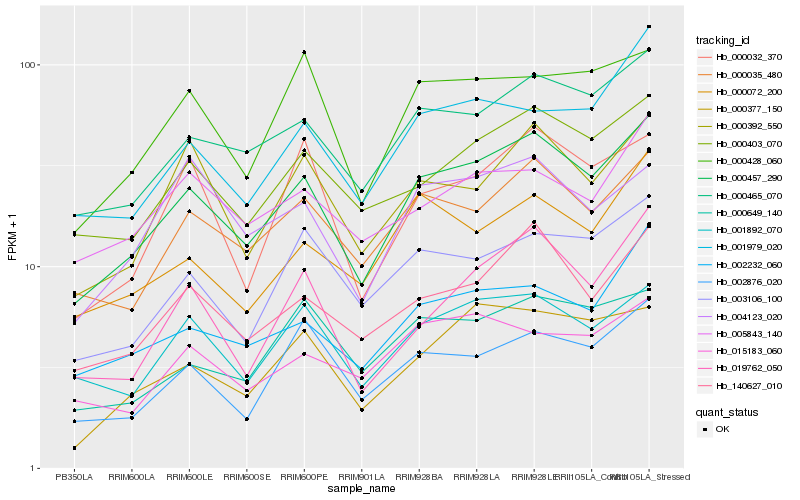
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_290 |
0.0 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 2 |
Hb_000465_070 |
0.0716817347 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase small subunit 3-like [Jatropha curcas] |
| 3 |
Hb_000035_480 |
0.0820320506 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 4 |
Hb_140627_010 |
0.083885305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000403_070 |
0.0856609965 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001892_070 |
0.0909681091 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 7 |
Hb_002232_060 |
0.0991539586 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 8 |
Hb_003106_100 |
0.1016701618 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000392_550 |
0.1023461922 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000377_150 |
0.1058007925 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 11 |
Hb_002876_020 |
0.1067193164 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000428_060 |
0.1075443354 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_019762_050 |
0.1085779443 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 14 |
Hb_000072_200 |
0.108932455 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 15 |
Hb_001979_020 |
0.108991937 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 16 |
Hb_000649_140 |
0.1115507836 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000032_370 |
0.1137563842 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 18 |
Hb_005843_140 |
0.1151225645 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_015183_060 |
0.11601408 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 20 |
Hb_004123_020 |
0.1170241832 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |