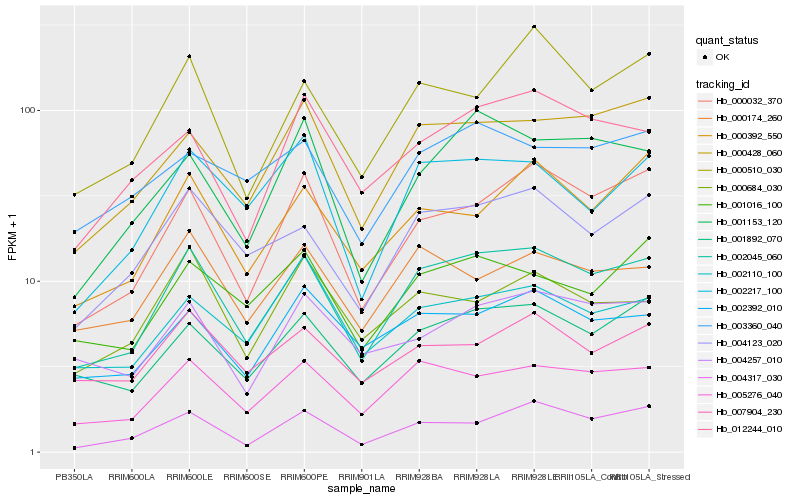
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_060 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_005276_040 |
0.0746980964 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 3 |
Hb_001892_070 |
0.0895843569 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 4 |
Hb_004123_020 |
0.0920996489 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000032_370 |
0.0926194326 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 6 |
Hb_000428_060 |
0.0956244049 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_004317_030 |
0.1052492287 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 8 |
Hb_002217_100 |
0.1081476042 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_002392_010 |
0.1106155135 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 10 |
Hb_000174_260 |
0.1115358046 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 11 |
Hb_001016_100 |
0.1127853294 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 12 |
Hb_000510_030 |
0.1143489807 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 13 |
Hb_012244_010 |
0.1152411889 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 14 |
Hb_007904_230 |
0.1155784755 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000392_550 |
0.1157826495 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001153_120 |
0.1168846457 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 17 |
Hb_004257_010 |
0.1171522025 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 18 |
Hb_002110_100 |
0.1210453165 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003360_040 |
0.121626317 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 20 |
Hb_000684_030 |
0.1243614923 |
- |
- |
syntaxin, putative [Ricinus communis] |