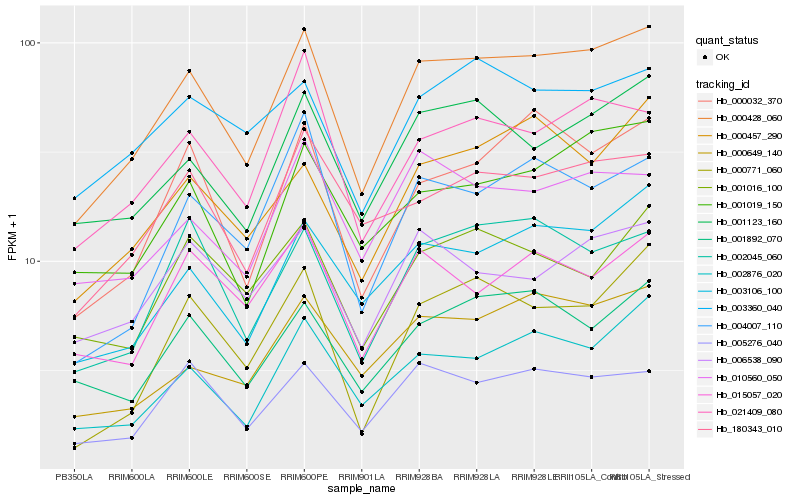
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000428_060 |
0.0 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_003106_100 |
0.088645667 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001123_160 |
0.0891253757 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 4 |
Hb_005276_040 |
0.0899764079 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 5 |
Hb_002876_020 |
0.0903023327 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 6 |
Hb_180343_010 |
0.0953192645 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 7 |
Hb_002045_060 |
0.0956244049 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001019_150 |
0.0975364961 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 9 |
Hb_000032_370 |
0.0976419219 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 10 |
Hb_001892_070 |
0.098580742 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 11 |
Hb_000649_140 |
0.0988306084 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_021409_080 |
0.1006423261 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 13 |
Hb_010560_050 |
0.1047111623 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 14 |
Hb_006538_090 |
0.1059039741 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 15 |
Hb_003360_040 |
0.1065393266 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 16 |
Hb_015057_020 |
0.1075229251 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 17 |
Hb_000457_290 |
0.1075443354 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 18 |
Hb_000771_060 |
0.1077351171 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 19 |
Hb_001016_100 |
0.1077668241 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 20 |
Hb_004007_110 |
0.1086164774 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |