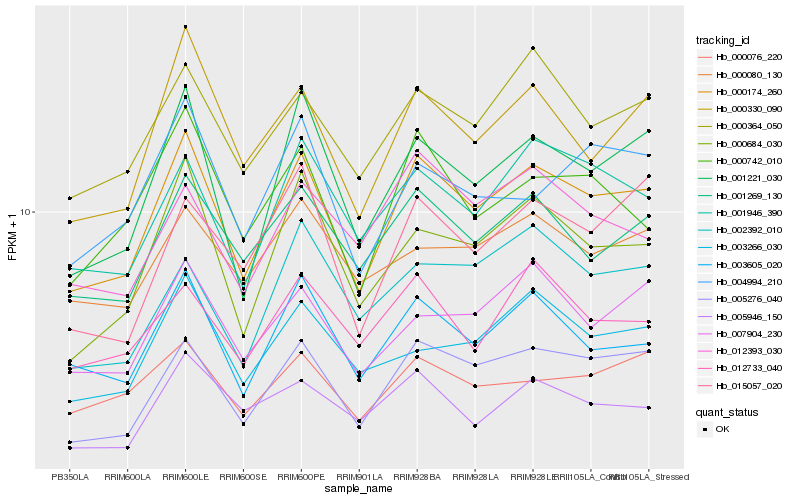
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_260 |
0.0 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 2 |
Hb_005276_040 |
0.0650859371 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 3 |
Hb_001221_030 |
0.074679828 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 4 |
Hb_003605_020 |
0.0748074179 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 5 |
Hb_000330_090 |
0.0756486883 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000684_030 |
0.0784204517 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_001269_130 |
0.0807049426 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 8 |
Hb_000364_050 |
0.0824484113 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001946_390 |
0.0847922391 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 10 |
Hb_007904_230 |
0.0886071098 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000742_010 |
0.0903307281 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_000076_220 |
0.0929649304 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003266_030 |
0.0941332781 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 14 |
Hb_005946_150 |
0.0974073131 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 15 |
Hb_004994_210 |
0.0981807798 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 16 |
Hb_012733_040 |
0.0989838172 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 17 |
Hb_012393_030 |
0.1009432826 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 18 |
Hb_000080_130 |
0.1018203407 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 19 |
Hb_015057_020 |
0.1023987958 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 20 |
Hb_002392_010 |
0.1038253085 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |