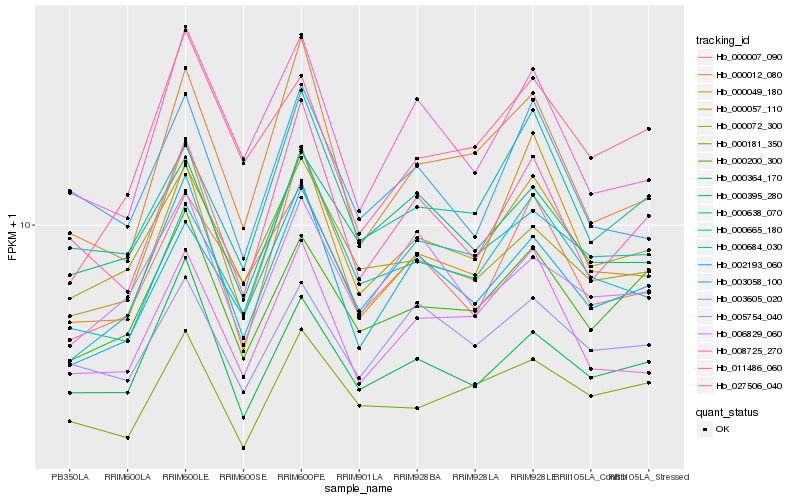
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_110 |
0.0 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 2 |
Hb_000684_030 |
0.076094111 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 3 |
Hb_000200_300 |
0.0791477545 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 4 |
Hb_000364_170 |
0.0805558137 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 5 |
Hb_000012_080 |
0.0864153282 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 6 |
Hb_006829_060 |
0.0888602096 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008725_270 |
0.0905463637 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 8 |
Hb_000007_090 |
0.090976244 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 9 |
Hb_002193_060 |
0.092501176 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 10 |
Hb_000181_350 |
0.0942248556 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 11 |
Hb_011486_060 |
0.0973612423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000665_180 |
0.0986943758 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_000638_070 |
0.1009190327 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 14 |
Hb_003605_020 |
0.1023743821 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 15 |
Hb_000049_180 |
0.1024521314 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 16 |
Hb_000072_300 |
0.1040204914 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 17 |
Hb_027506_040 |
0.1043604477 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005754_040 |
0.1046712019 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003058_100 |
0.1053078533 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000395_280 |
0.1057517943 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |