| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
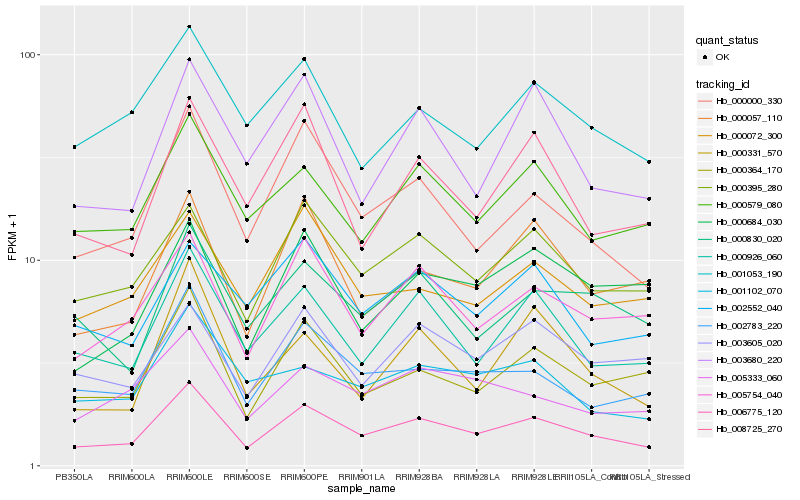
Hb_006775_120 |
0.0 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 2 |
Hb_000000_330 |
0.0892824509 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 3 |
Hb_000926_060 |
0.0973259004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005754_040 |
0.0975802018 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000364_170 |
0.0989397978 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 6 |
Hb_000395_280 |
0.0999453288 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 7 |
Hb_002783_220 |
0.1022304062 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000579_080 |
0.1079413445 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 9 |
Hb_000057_110 |
0.1103025467 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 10 |
Hb_008725_270 |
0.1136084574 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 11 |
Hb_003680_220 |
0.1146569469 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 12 |
Hb_000830_020 |
0.1158884143 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 13 |
Hb_005333_060 |
0.1180687482 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 14 |
Hb_003605_020 |
0.1188749232 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 15 |
Hb_001053_190 |
0.1202018012 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 16 |
Hb_000684_030 |
0.1208130785 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_001102_070 |
0.1215765569 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000331_570 |
0.1216317985 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 19 |
Hb_002552_040 |
0.1230700296 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 20 |
Hb_000072_300 |
0.1235673475 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |