| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
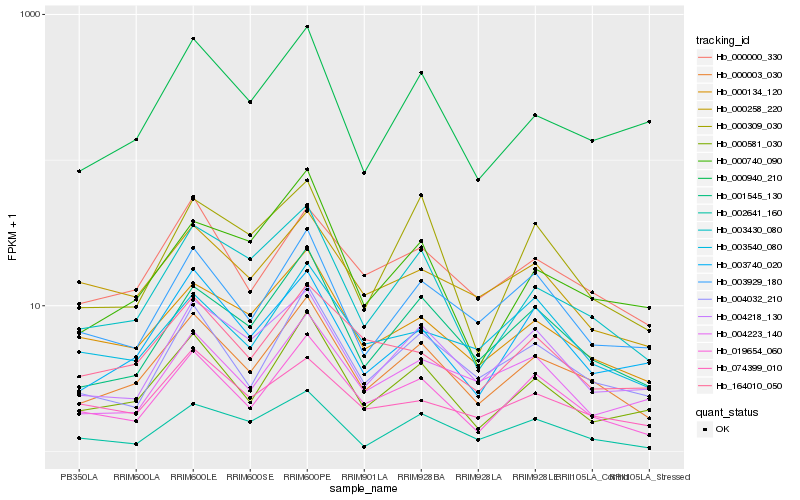
Hb_000003_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001545_130 |
0.0902195768 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 3 |
Hb_019654_060 |
0.0930110222 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003430_080 |
0.0957894033 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_004032_210 |
0.1018278102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000581_030 |
0.1046411475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000000_330 |
0.1052534439 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 8 |
Hb_000134_120 |
0.1053576604 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 9 |
Hb_003929_180 |
0.1107422905 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 10 |
Hb_004218_130 |
0.1117993473 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 11 |
Hb_000940_210 |
0.1149537463 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 12 |
Hb_003740_020 |
0.1163283763 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000258_220 |
0.1308165416 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_164010_050 |
0.1316193808 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 15 |
Hb_000740_090 |
0.1335624292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002641_160 |
0.1344695152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004223_140 |
0.1352733005 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 18 |
Hb_074399_010 |
0.1362616881 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 19 |
Hb_003540_080 |
0.1362921503 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000309_030 |
0.1375692939 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |