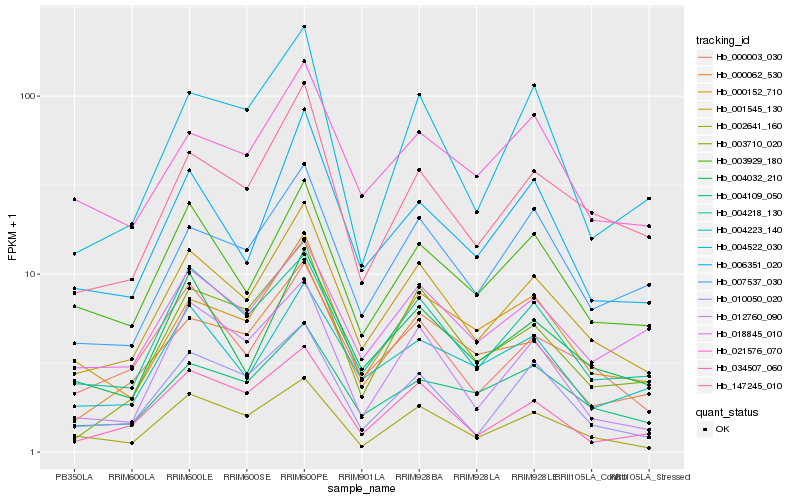
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001545_130 |
0.0 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 2 |
Hb_004218_130 |
0.0860799983 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 3 |
Hb_147245_010 |
0.0867830161 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 4 |
Hb_000003_030 |
0.0902195768 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_004109_050 |
0.0926537536 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 6 |
Hb_006351_020 |
0.0961901446 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 7 |
Hb_004032_210 |
0.0984217505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003929_180 |
0.0995211453 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 9 |
Hb_000152_710 |
0.1005915761 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 10 |
Hb_000062_530 |
0.1006071585 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 11 |
Hb_007537_030 |
0.1045681091 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002641_160 |
0.1077514359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_021576_070 |
0.1096143929 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 14 |
Hb_004223_140 |
0.1101348432 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_018845_010 |
0.1102062651 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 16 |
Hb_010050_020 |
0.1110725707 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 17 |
Hb_003710_020 |
0.1125331118 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 18 |
Hb_034507_060 |
0.1152726177 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 19 |
Hb_012760_090 |
0.1160705933 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 20 |
Hb_004522_030 |
0.117884838 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |