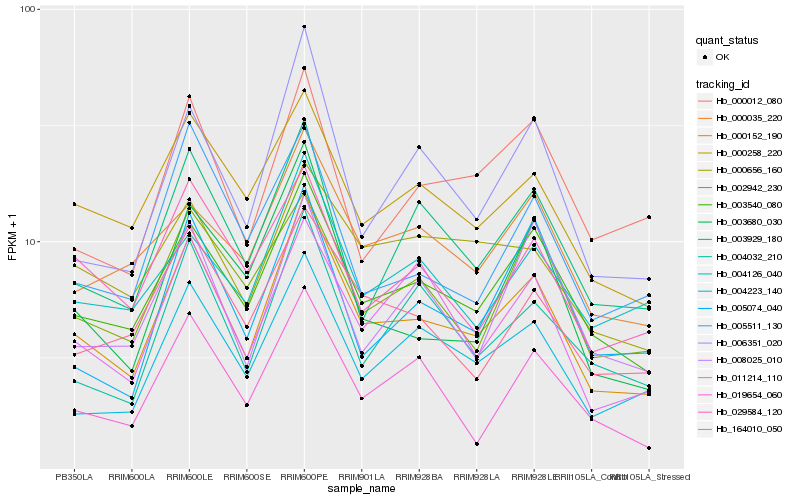
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_190 |
0.0 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 2 |
Hb_003540_080 |
0.0808072108 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000258_220 |
0.107507125 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006351_020 |
0.1077766833 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 5 |
Hb_004223_140 |
0.1084183858 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 6 |
Hb_003929_180 |
0.1127831995 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 7 |
Hb_164010_050 |
0.1150149138 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 8 |
Hb_011214_110 |
0.1160629055 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 9 |
Hb_004032_210 |
0.118780318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_029584_120 |
0.12000389 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_019654_060 |
0.120736215 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002942_230 |
0.1208503408 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 13 |
Hb_003680_030 |
0.1217507797 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000035_220 |
0.1218029433 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 15 |
Hb_005511_130 |
0.1272262673 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 16 |
Hb_000012_080 |
0.1284578647 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 17 |
Hb_004126_040 |
0.1285082508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000656_160 |
0.1289206169 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 19 |
Hb_005074_040 |
0.1290619424 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_008025_010 |
0.1299197899 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |