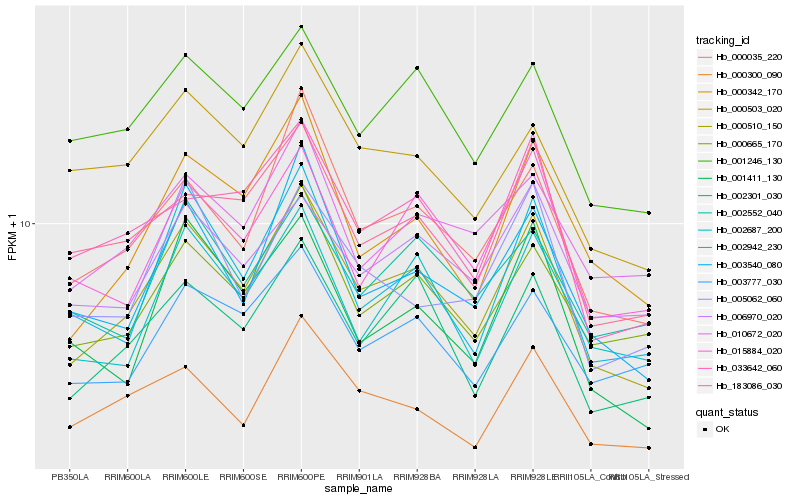
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_150 |
0.0 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002301_030 |
0.0767406098 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 3 |
Hb_000035_220 |
0.0770747507 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 4 |
Hb_000342_170 |
0.0857629783 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 5 |
Hb_000665_170 |
0.0863920587 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 6 |
Hb_003540_080 |
0.0873389641 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002942_230 |
0.0876296908 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 8 |
Hb_002687_200 |
0.0919776164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_183086_030 |
0.0921828 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 10 |
Hb_001246_130 |
0.0954550997 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000300_090 |
0.0955446789 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_015884_020 |
0.0956847088 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 13 |
Hb_033642_060 |
0.0963449595 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003777_030 |
0.0981716311 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 15 |
Hb_006970_020 |
0.1010038628 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 16 |
Hb_010672_020 |
0.1035455315 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001411_130 |
0.1038160819 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000503_020 |
0.1040687347 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 19 |
Hb_005062_060 |
0.1050653288 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 20 |
Hb_002552_040 |
0.1068761185 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |