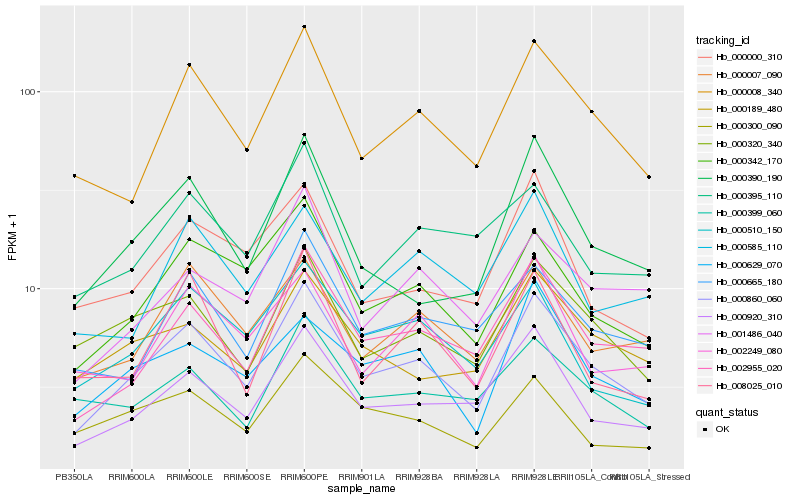
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000860_060 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 2 |
Hb_002955_020 |
0.0893331042 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_000920_310 |
0.0920551793 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_000008_340 |
0.0921682742 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 5 |
Hb_000342_170 |
0.0949456616 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 6 |
Hb_000320_340 |
0.1085628233 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 7 |
Hb_000390_190 |
0.108661945 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000007_090 |
0.1115723503 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 9 |
Hb_000665_180 |
0.1134958433 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_000629_070 |
0.1174089655 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 11 |
Hb_000510_150 |
0.1191877779 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001486_040 |
0.1192511836 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000189_480 |
0.1195232473 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 14 |
Hb_002249_080 |
0.1223205493 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 15 |
Hb_000000_310 |
0.1232906545 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 16 |
Hb_000585_110 |
0.1245739346 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 17 |
Hb_000399_060 |
0.1257311135 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 18 |
Hb_000395_110 |
0.125805322 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 19 |
Hb_008025_010 |
0.1259942901 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 20 |
Hb_000300_090 |
0.1263607943 |
- |
- |
catalytic, putative [Ricinus communis] |