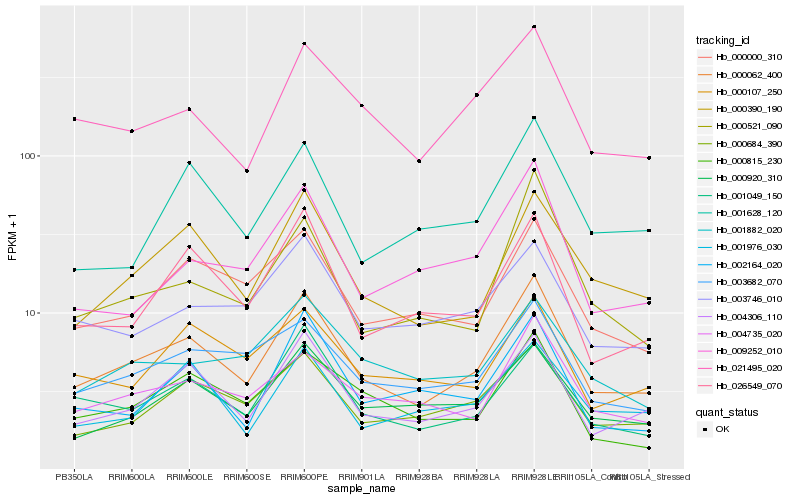
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_400 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004306_110 |
0.0890785866 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 3 |
Hb_000390_190 |
0.1010769542 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000684_390 |
0.1091875169 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 5 |
Hb_004735_020 |
0.1133456134 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 6 |
Hb_021495_020 |
0.1135042629 |
- |
- |
chitinase, putative [Ricinus communis] |
| 7 |
Hb_001049_150 |
0.1151216205 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 8 |
Hb_000920_310 |
0.1157422524 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_026549_070 |
0.1164224844 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 10 |
Hb_000000_310 |
0.1168589936 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_003682_070 |
0.1191282999 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000815_230 |
0.1196417143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001628_120 |
0.1262212008 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 14 |
Hb_000521_090 |
0.1267857546 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009252_010 |
0.128554987 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 16 |
Hb_002164_020 |
0.1292330562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001882_020 |
0.1298598819 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 18 |
Hb_003746_010 |
0.1302155059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000107_250 |
0.1311505735 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 20 |
Hb_001976_030 |
0.1327300939 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |