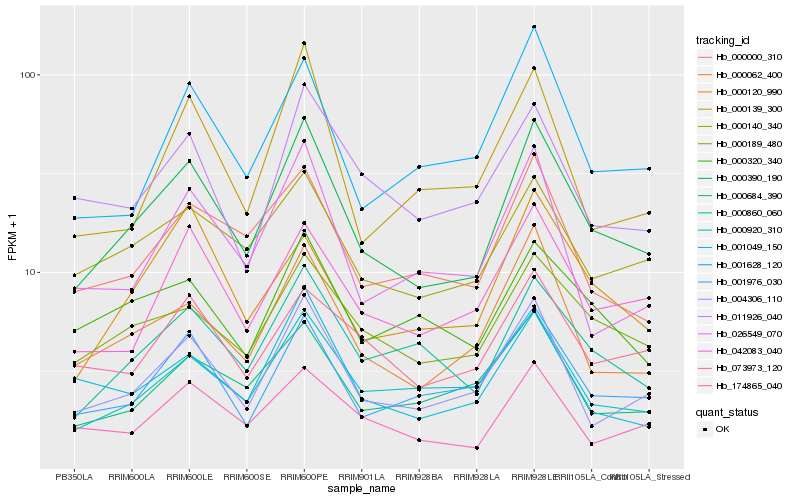
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_190 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_004306_110 |
0.098254192 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 3 |
Hb_000062_400 |
0.1010769542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000860_060 |
0.108661945 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 5 |
Hb_000189_480 |
0.1116357938 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 6 |
Hb_001976_030 |
0.1121217311 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000920_310 |
0.1157586353 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_042083_040 |
0.1172544898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000140_340 |
0.1187823778 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 10 |
Hb_000000_310 |
0.1188484314 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_073973_120 |
0.1211762462 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001628_120 |
0.1241824756 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 13 |
Hb_000684_390 |
0.1261047593 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 14 |
Hb_026549_070 |
0.1269593515 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 15 |
Hb_174865_040 |
0.1288370966 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001049_150 |
0.1304035262 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 17 |
Hb_000139_300 |
0.1304499235 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 18 |
Hb_000320_340 |
0.1305088336 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 19 |
Hb_011926_040 |
0.1323032587 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000120_990 |
0.1331740476 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |