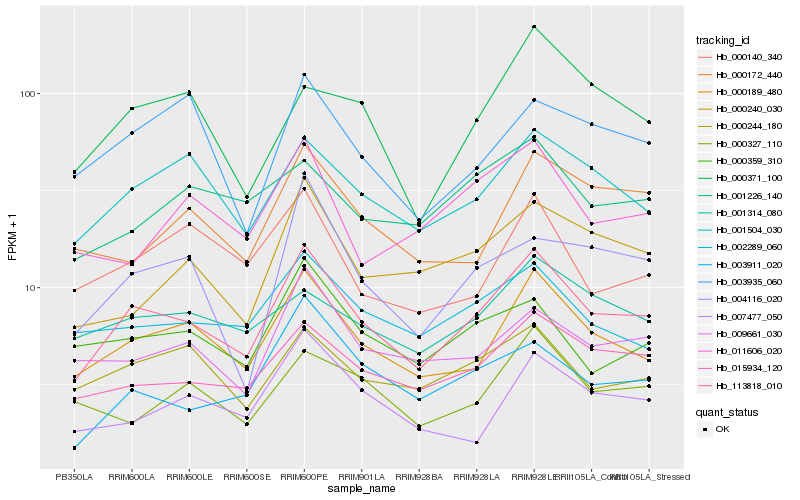
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113818_010 |
0.0 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 2 |
Hb_000189_480 |
0.0871936732 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 3 |
Hb_015934_120 |
0.1004853016 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 4 |
Hb_001504_030 |
0.1105869005 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 5 |
Hb_002289_060 |
0.1215454706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000244_180 |
0.1226098622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009661_030 |
0.1229414946 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_000240_030 |
0.1255011463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003911_020 |
0.1272911567 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 10 |
Hb_001314_080 |
0.1297015639 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 11 |
Hb_000359_310 |
0.1304773183 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 12 |
Hb_000172_440 |
0.1309068415 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 13 |
Hb_001226_140 |
0.1364868429 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 14 |
Hb_000327_110 |
0.1370644393 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 15 |
Hb_007477_050 |
0.1370739019 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000140_340 |
0.1371537578 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 17 |
Hb_004116_020 |
0.1374782764 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 18 |
Hb_000371_100 |
0.1386163712 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 19 |
Hb_003935_060 |
0.1386804403 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 20 |
Hb_011606_020 |
0.1391285713 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |