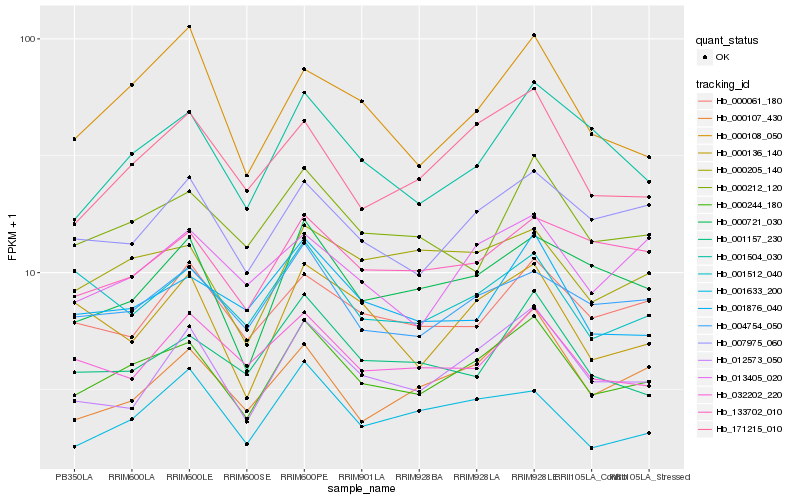
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000244_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012573_050 |
0.0825292879 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 3 |
Hb_004754_050 |
0.0843324372 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 4 |
Hb_171215_010 |
0.0860146972 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 5 |
Hb_007975_060 |
0.0884393892 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000721_030 |
0.0898838918 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 7 |
Hb_001876_040 |
0.0899463628 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 8 |
Hb_000205_140 |
0.0903438467 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 9 |
Hb_001504_030 |
0.0911964308 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 10 |
Hb_000212_120 |
0.0925134891 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 11 |
Hb_133702_010 |
0.0962540906 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_001157_230 |
0.0967560743 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 13 |
Hb_000061_180 |
0.0991614321 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 14 |
Hb_000107_430 |
0.1003187433 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 15 |
Hb_001512_040 |
0.1004731829 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 16 |
Hb_001633_200 |
0.1006667478 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 17 |
Hb_032202_220 |
0.1011467916 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_013405_020 |
0.1011513207 |
- |
- |
- |
| 19 |
Hb_000136_140 |
0.1015743845 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 20 |
Hb_000108_050 |
0.101922312 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |