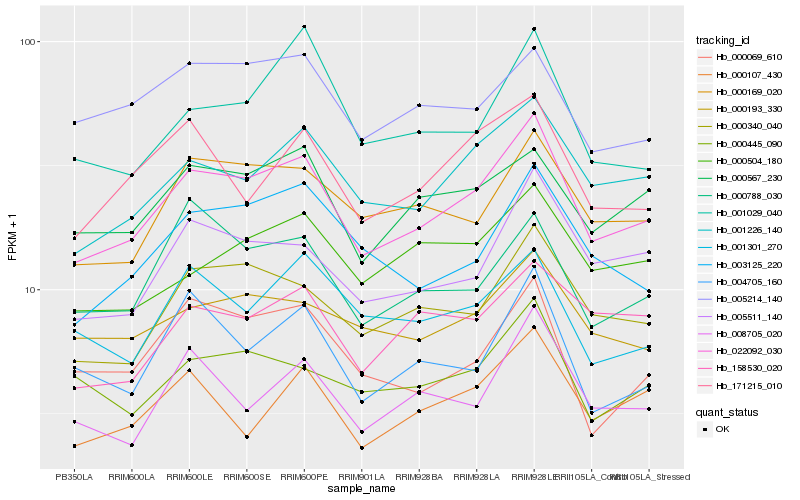
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_030 |
0.0 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001226_140 |
0.0721525295 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 3 |
Hb_000340_040 |
0.0764722632 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 4 |
Hb_005511_140 |
0.0803625039 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 5 |
Hb_000169_020 |
0.0812409185 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 6 |
Hb_000567_230 |
0.083749514 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 7 |
Hb_003125_220 |
0.088734293 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 8 |
Hb_000193_330 |
0.0894250275 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 9 |
Hb_171215_010 |
0.0933508091 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 10 |
Hb_000788_030 |
0.0942981628 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 11 |
Hb_000445_090 |
0.0946468788 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 12 |
Hb_000504_180 |
0.0949346809 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_005214_140 |
0.0953389953 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 14 |
Hb_000069_610 |
0.0964326894 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 15 |
Hb_008705_020 |
0.0968944743 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000107_430 |
0.096951418 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 17 |
Hb_001301_270 |
0.0971015412 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 18 |
Hb_158530_020 |
0.0985162779 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 19 |
Hb_004705_160 |
0.0985296282 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 20 |
Hb_001029_040 |
0.0990545936 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |