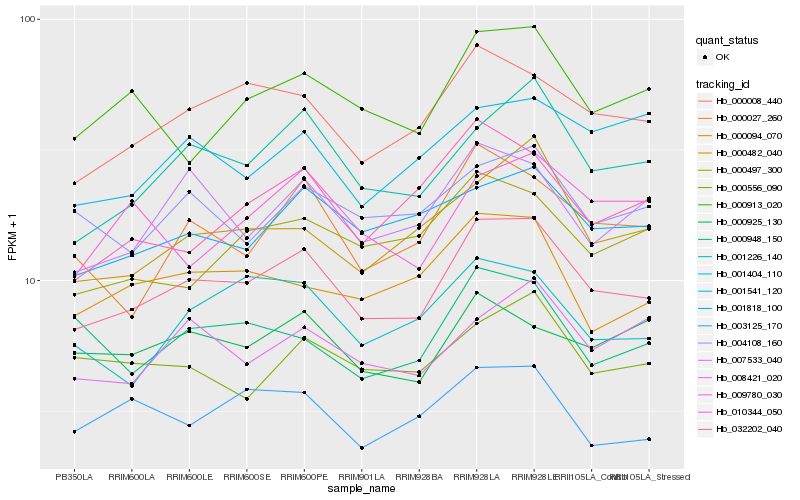
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_040 |
0.0 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 2 |
Hb_000008_440 |
0.0698455347 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Jatropha curcas] |
| 3 |
Hb_001226_140 |
0.0705528285 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 4 |
Hb_001818_100 |
0.0836428002 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 5 |
Hb_007533_040 |
0.0840380907 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 6 |
Hb_000482_040 |
0.0848736365 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 7 |
Hb_000027_260 |
0.0864978928 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 8 |
Hb_000913_020 |
0.0891337408 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001404_110 |
0.0904654131 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_000094_070 |
0.0914606937 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004108_160 |
0.0920907828 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 12 |
Hb_000497_300 |
0.0933460727 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 13 |
Hb_008421_020 |
0.0936011414 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 14 |
Hb_010344_050 |
0.096658762 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 15 |
Hb_000925_130 |
0.0967404501 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_003125_170 |
0.0969457783 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 17 |
Hb_000556_090 |
0.0975893242 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001541_120 |
0.0989778717 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 19 |
Hb_009780_030 |
0.100404506 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 20 |
Hb_000948_150 |
0.1007312031 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |