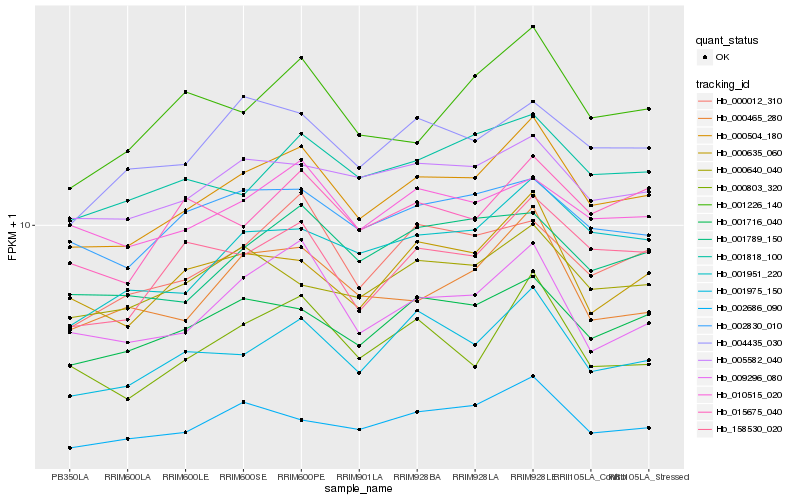
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_180 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 2 |
Hb_001716_040 |
0.0649624139 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 3 |
Hb_001975_150 |
0.0670548168 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 4 |
Hb_158530_020 |
0.0716894265 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 5 |
Hb_001951_220 |
0.0723171033 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 6 |
Hb_009296_080 |
0.075046617 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005582_040 |
0.0751130205 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 8 |
Hb_002686_090 |
0.0756347012 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 9 |
Hb_000635_060 |
0.0756720448 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 10 |
Hb_000465_280 |
0.076638395 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 11 |
Hb_001789_150 |
0.0779270858 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001818_100 |
0.0781581599 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 13 |
Hb_000012_310 |
0.078631117 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_002830_010 |
0.0786553803 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_004435_030 |
0.0809872721 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 16 |
Hb_000803_320 |
0.0814095141 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 17 |
Hb_010515_020 |
0.0816125744 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 18 |
Hb_015675_040 |
0.0817631142 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 19 |
Hb_000640_040 |
0.082284075 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001226_140 |
0.0824154315 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |