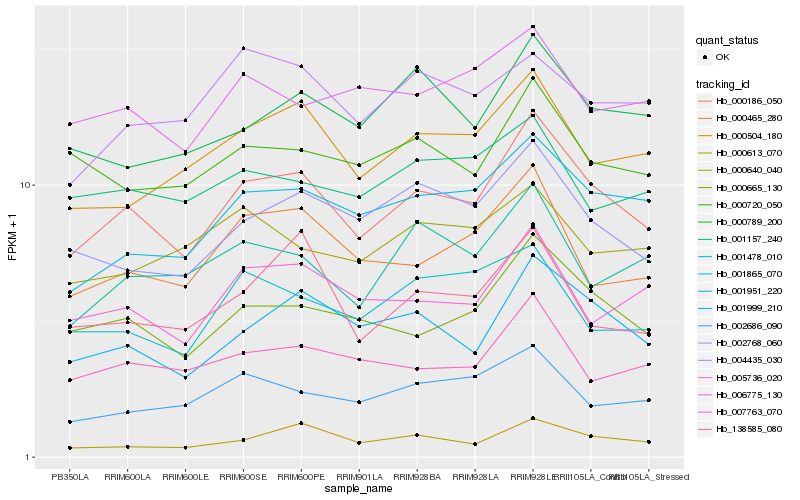
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 2 |
Hb_000665_130 |
0.0728809041 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001999_210 |
0.0785799393 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 4 |
Hb_001951_220 |
0.0845183454 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 5 |
Hb_000504_180 |
0.089906651 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_002768_060 |
0.0903969614 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000465_280 |
0.0915459765 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 8 |
Hb_006775_130 |
0.0936757454 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 9 |
Hb_007763_070 |
0.0967048934 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000720_050 |
0.0982091616 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000789_200 |
0.0982614603 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 12 |
Hb_001478_010 |
0.0988189696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002686_090 |
0.1024315201 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 14 |
Hb_138585_080 |
0.1045081469 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 15 |
Hb_005736_020 |
0.1045291986 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 16 |
Hb_000613_070 |
0.1061074703 |
- |
- |
- |
| 17 |
Hb_001865_070 |
0.1063138529 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001157_240 |
0.1063944654 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 19 |
Hb_004435_030 |
0.1065265439 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 20 |
Hb_000640_040 |
0.1079024928 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |