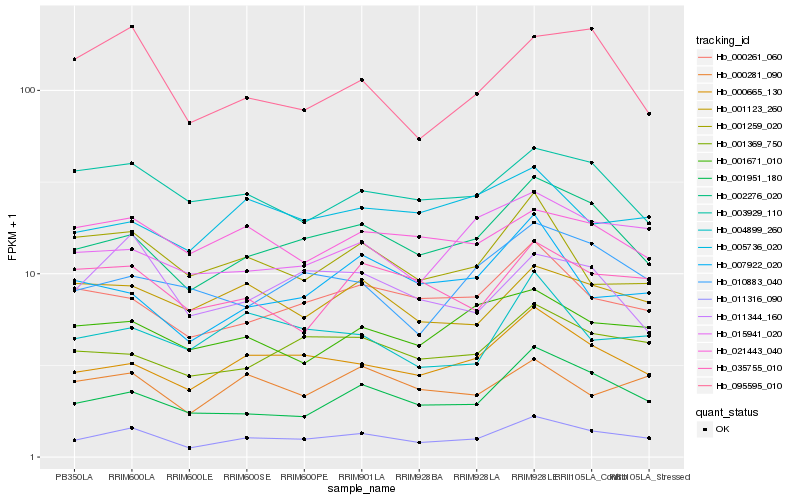
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011316_090 |
0.0 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 2 |
Hb_002276_020 |
0.0750395165 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000665_130 |
0.0886213336 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001951_180 |
0.0959139898 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000261_060 |
0.1038173392 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 6 |
Hb_001369_750 |
0.1040546492 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 7 |
Hb_000281_090 |
0.1194425734 |
transcription factor |
TF Family: SNF2 |
hypothetical protein PRUPE_ppa000063mg [Prunus persica] |
| 8 |
Hb_015941_020 |
0.1196558672 |
- |
- |
- |
| 9 |
Hb_004899_260 |
0.1197734446 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 10 |
Hb_001671_010 |
0.1204247101 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 11 |
Hb_003929_110 |
0.1204622554 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001259_020 |
0.1223667602 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 13 |
Hb_035755_010 |
0.1225398869 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_001123_260 |
0.1232566957 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 15 |
Hb_010883_040 |
0.1233248468 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 16 |
Hb_007922_020 |
0.1239113424 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 17 |
Hb_095595_010 |
0.1255566315 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 18 |
Hb_021443_040 |
0.1266505763 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 19 |
Hb_011344_160 |
0.1267552048 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 20 |
Hb_005736_020 |
0.1268298715 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |