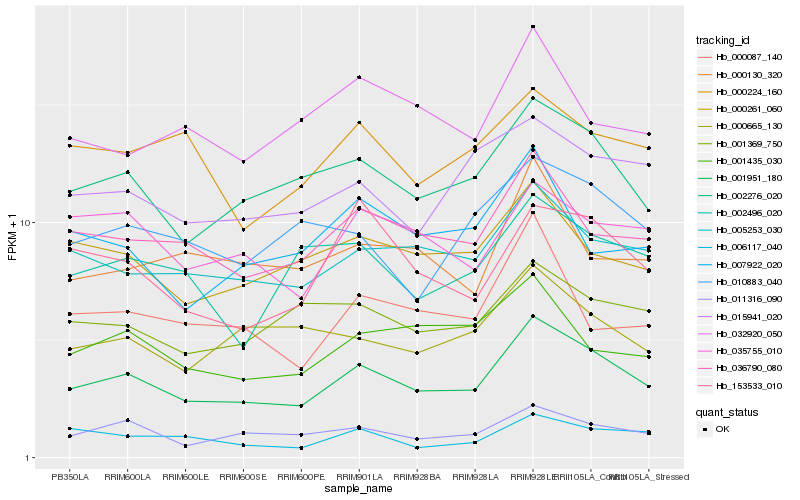
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_180 |
0.0 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_002276_020 |
0.0826194562 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 3 |
Hb_036790_080 |
0.094909099 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 4 |
Hb_011316_090 |
0.0959139898 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 5 |
Hb_005253_030 |
0.097578981 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000261_060 |
0.1086221991 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 7 |
Hb_153533_010 |
0.1121136713 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 8 |
Hb_001369_750 |
0.1136988938 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 9 |
Hb_035755_010 |
0.1139997872 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_015941_020 |
0.1160708906 |
- |
- |
- |
| 11 |
Hb_001435_030 |
0.1183941532 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 12 |
Hb_000665_130 |
0.1187525017 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000087_140 |
0.1202273605 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002496_020 |
0.1205344637 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_006117_040 |
0.1205610881 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 16 |
Hb_010883_040 |
0.1216093742 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 17 |
Hb_000224_160 |
0.1225536479 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_000130_320 |
0.1235980616 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 19 |
Hb_007922_020 |
0.1246334621 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 20 |
Hb_032920_050 |
0.1259193892 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |