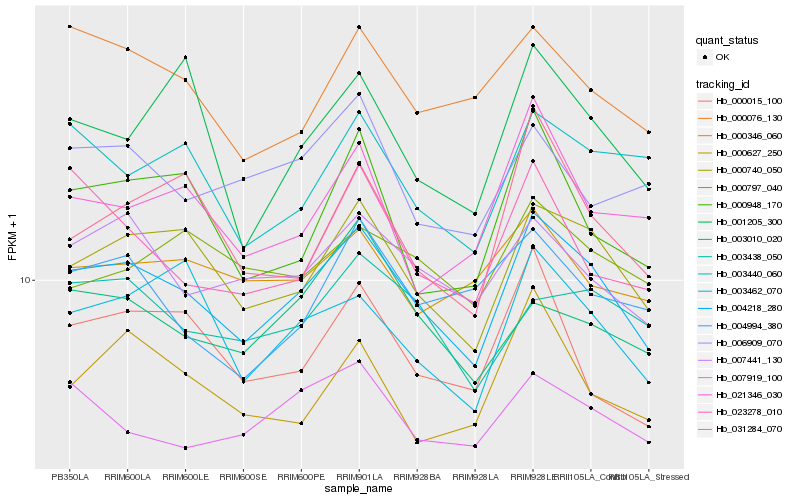
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_280 |
0.0 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 2 |
Hb_000740_050 |
0.059367651 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 3 |
Hb_007441_130 |
0.0843925027 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003462_070 |
0.0899320385 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 5 |
Hb_000076_130 |
0.0933324103 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 6 |
Hb_007919_100 |
0.0950805062 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 7 |
Hb_003438_050 |
0.095281288 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 8 |
Hb_003440_060 |
0.0975022884 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 9 |
Hb_000948_170 |
0.0982478831 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 10 |
Hb_000346_060 |
0.0993873772 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 11 |
Hb_031284_070 |
0.1002795192 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000627_250 |
0.1005110694 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 13 |
Hb_023278_010 |
0.1008832614 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 14 |
Hb_000015_100 |
0.1009724726 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_006909_070 |
0.1022368867 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_021346_030 |
0.1023441271 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 17 |
Hb_001205_300 |
0.1025370361 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 18 |
Hb_003010_020 |
0.1026936963 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000797_040 |
0.1027483506 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 20 |
Hb_004994_380 |
0.1046271717 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |