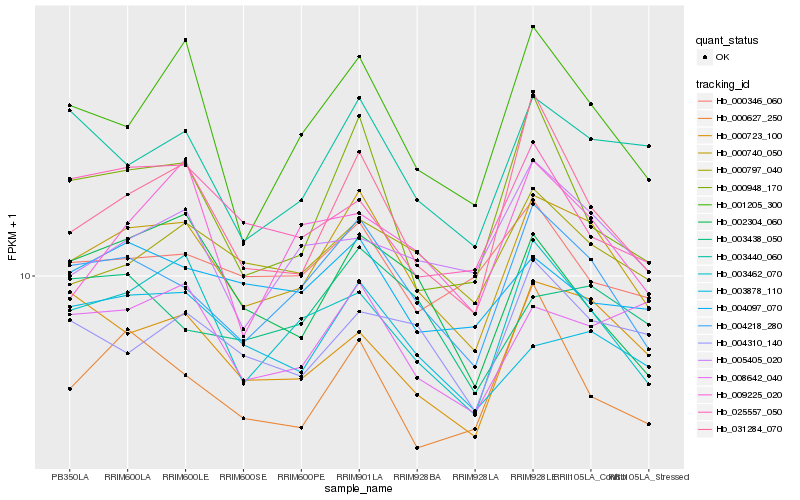
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_050 |
0.0 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 2 |
Hb_004218_280 |
0.059367651 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 3 |
Hb_003462_070 |
0.0753947149 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 4 |
Hb_031284_070 |
0.0852988795 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001205_300 |
0.08763927 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 6 |
Hb_000797_040 |
0.0881472634 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 7 |
Hb_003440_060 |
0.0884454977 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 8 |
Hb_000723_100 |
0.0942542861 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 9 |
Hb_000948_170 |
0.0957216503 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 10 |
Hb_008642_040 |
0.0960503644 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003878_110 |
0.0980048116 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 12 |
Hb_004097_070 |
0.0981376089 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 13 |
Hb_004310_140 |
0.0984697162 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002304_060 |
0.0990212323 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 15 |
Hb_003438_050 |
0.0991647047 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 16 |
Hb_000346_060 |
0.1015112676 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 17 |
Hb_005405_020 |
0.1022127179 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_000627_250 |
0.1022880446 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 19 |
Hb_025557_050 |
0.1033962987 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 20 |
Hb_009225_020 |
0.103605684 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |