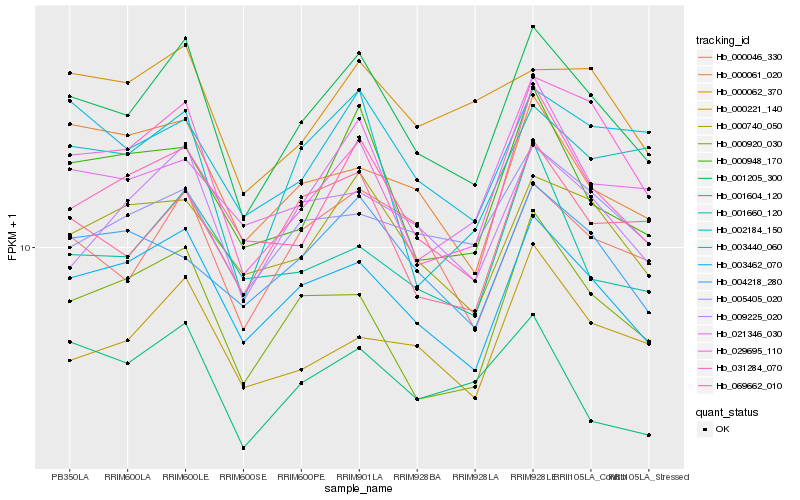
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_300 |
0.0 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 2 |
Hb_003462_070 |
0.0639538618 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_031284_070 |
0.0864331724 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000046_330 |
0.0868595779 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 5 |
Hb_000740_050 |
0.08763927 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 6 |
Hb_009225_020 |
0.0876609167 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 7 |
Hb_000948_170 |
0.0912107027 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 8 |
Hb_029695_110 |
0.0915571399 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 9 |
Hb_069662_010 |
0.0929657062 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 10 |
Hb_000920_030 |
0.0935228168 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_003440_060 |
0.0996150988 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 12 |
Hb_000061_020 |
0.1000422263 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 13 |
Hb_021346_030 |
0.1010636532 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 14 |
Hb_004218_280 |
0.1025370361 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 15 |
Hb_000221_140 |
0.1030082503 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_005405_020 |
0.103251894 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_002184_150 |
0.1057870616 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000062_370 |
0.1059566222 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 19 |
Hb_001660_120 |
0.1062482045 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001604_120 |
0.1063108321 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |