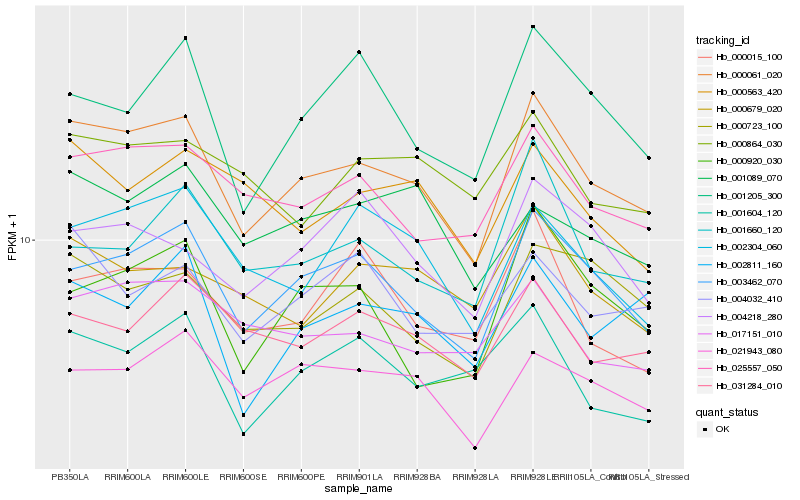
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 2 |
Hb_003462_070 |
0.0653356846 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_025557_050 |
0.0826451312 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 4 |
Hb_021943_080 |
0.0911045419 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000723_100 |
0.0971719314 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 6 |
Hb_002304_060 |
0.0977984736 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 7 |
Hb_017151_010 |
0.0983654386 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 8 |
Hb_031284_010 |
0.0984430559 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 9 |
Hb_001205_300 |
0.1000422263 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 10 |
Hb_002811_160 |
0.1010808647 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000679_020 |
0.1025472602 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 12 |
Hb_001660_120 |
0.1032114704 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004032_410 |
0.1047783617 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000563_420 |
0.1055816808 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 15 |
Hb_001089_070 |
0.106014398 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_001604_120 |
0.1061682255 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000920_030 |
0.1063799165 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000864_030 |
0.1064833892 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004218_280 |
0.1082211193 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 20 |
Hb_000015_100 |
0.1090003417 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |