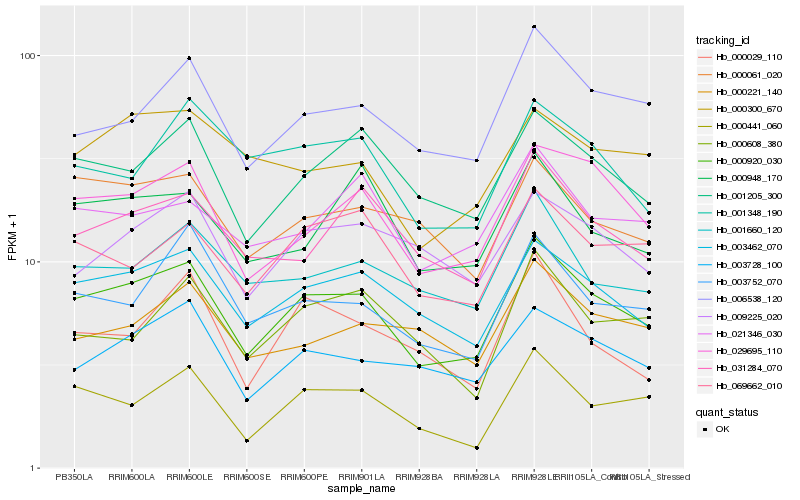
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_030 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_003462_070 |
0.0753338071 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_029695_110 |
0.086261163 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 4 |
Hb_001205_300 |
0.0935228168 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 5 |
Hb_006538_120 |
0.0951601454 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003728_100 |
0.1001655285 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000441_060 |
0.1023635705 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 8 |
Hb_001660_120 |
0.1033124571 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003752_070 |
0.1037520086 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031284_070 |
0.1058242786 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000061_020 |
0.1063799165 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 12 |
Hb_069662_010 |
0.1064443415 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 13 |
Hb_021346_030 |
0.1127872888 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 14 |
Hb_009225_020 |
0.1128538408 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 15 |
Hb_000948_170 |
0.1141894865 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 16 |
Hb_001348_190 |
0.114425962 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000221_140 |
0.1150263849 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000300_670 |
0.1150920444 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000608_380 |
0.1166466232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000029_110 |
0.1176869709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |