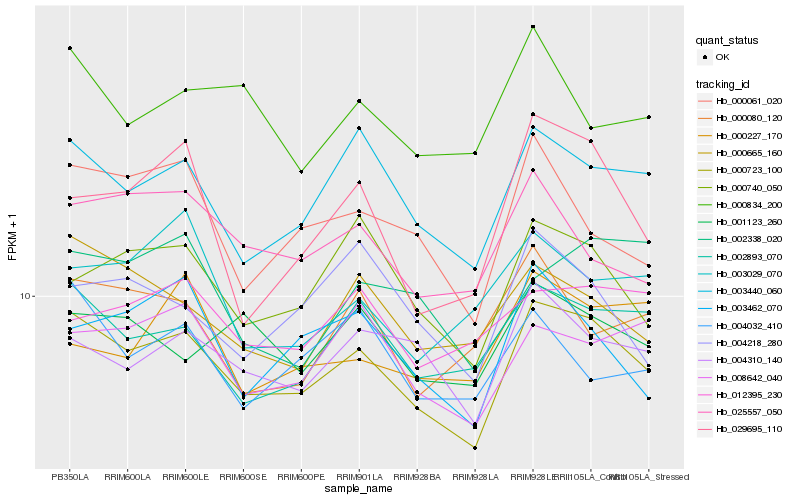
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_100 |
0.0 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 2 |
Hb_003440_060 |
0.0736846612 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 3 |
Hb_002893_070 |
0.0827801666 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004310_140 |
0.0891768589 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000665_160 |
0.0920130915 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000740_050 |
0.0942542861 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000061_020 |
0.0971719314 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 8 |
Hb_008642_040 |
0.0973660208 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_025557_050 |
0.0986445979 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 10 |
Hb_003462_070 |
0.1000068542 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 11 |
Hb_001123_260 |
0.1012111327 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 12 |
Hb_029695_110 |
0.1044972366 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 13 |
Hb_012395_230 |
0.1048539294 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000834_200 |
0.1056929327 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 15 |
Hb_000080_120 |
0.1064231255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 16 |
Hb_004218_280 |
0.1065314326 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 17 |
Hb_004032_410 |
0.1066121413 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002338_020 |
0.1090403589 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 19 |
Hb_003029_070 |
0.1118783936 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000227_170 |
0.1131824332 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |