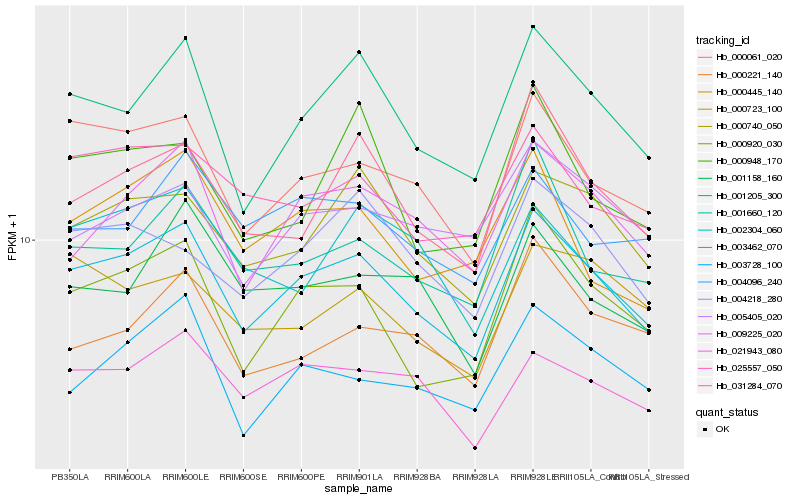
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003462_070 |
0.0 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 2 |
Hb_001205_300 |
0.0639538618 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 3 |
Hb_000061_020 |
0.0653356846 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 4 |
Hb_000920_030 |
0.0753338071 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000740_050 |
0.0753947149 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 6 |
Hb_009225_020 |
0.0770397768 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 7 |
Hb_021943_080 |
0.0811314001 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_025557_050 |
0.0858910845 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 9 |
Hb_031284_070 |
0.0864858351 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004218_280 |
0.0899320385 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 11 |
Hb_003728_100 |
0.0899330017 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001660_120 |
0.0908985665 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000445_140 |
0.0942610046 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 14 |
Hb_004096_240 |
0.094420324 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 15 |
Hb_001158_160 |
0.0962877035 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005405_020 |
0.0964915563 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_002304_060 |
0.0979470809 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 18 |
Hb_000221_140 |
0.0988968068 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000723_100 |
0.1000068542 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 20 |
Hb_000948_170 |
0.1003156041 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |