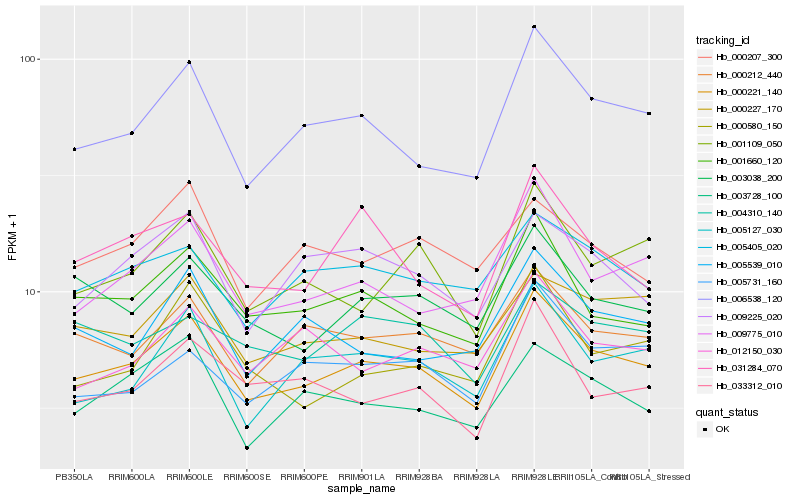
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_140 |
0.0 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_006538_120 |
0.0617143078 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001660_120 |
0.0753639263 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009775_010 |
0.077574298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 5 |
Hb_000212_440 |
0.0798990372 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_003038_200 |
0.082131872 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 7 |
Hb_005405_020 |
0.0845617381 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_005127_030 |
0.0849038582 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_003728_100 |
0.0866007668 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001109_050 |
0.0867301214 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 11 |
Hb_000227_170 |
0.0883661995 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_009225_020 |
0.0902492099 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 13 |
Hb_005539_010 |
0.0918828417 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 14 |
Hb_033312_010 |
0.0920266573 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_031284_070 |
0.092160893 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000580_150 |
0.0940212177 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005731_160 |
0.0948443818 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_012150_030 |
0.0955719137 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004310_140 |
0.0967160358 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_000207_300 |
0.0967467983 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |