| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
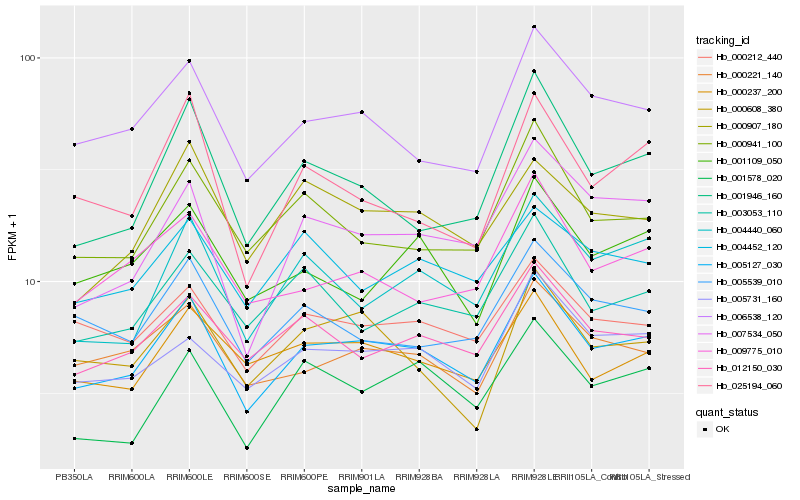
Hb_005127_030 |
0.0 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 2 |
Hb_006538_120 |
0.0746773962 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000221_140 |
0.0849038582 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_007534_050 |
0.0863632205 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_005731_160 |
0.0871429257 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_012150_030 |
0.0878790185 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000212_440 |
0.0885569469 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_004440_060 |
0.0893989547 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 9 |
Hb_001946_160 |
0.0894932041 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 10 |
Hb_000941_100 |
0.0895509111 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 11 |
Hb_000237_200 |
0.0931399879 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 12 |
Hb_003053_110 |
0.0945271829 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009775_010 |
0.094669156 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 14 |
Hb_000907_180 |
0.0953809192 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005539_010 |
0.0968237224 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 16 |
Hb_001578_020 |
0.0980399581 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 17 |
Hb_025194_060 |
0.0995861997 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_001109_050 |
0.1002769687 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 19 |
Hb_004452_120 |
0.1003299056 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 20 |
Hb_000608_380 |
0.1007102804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |