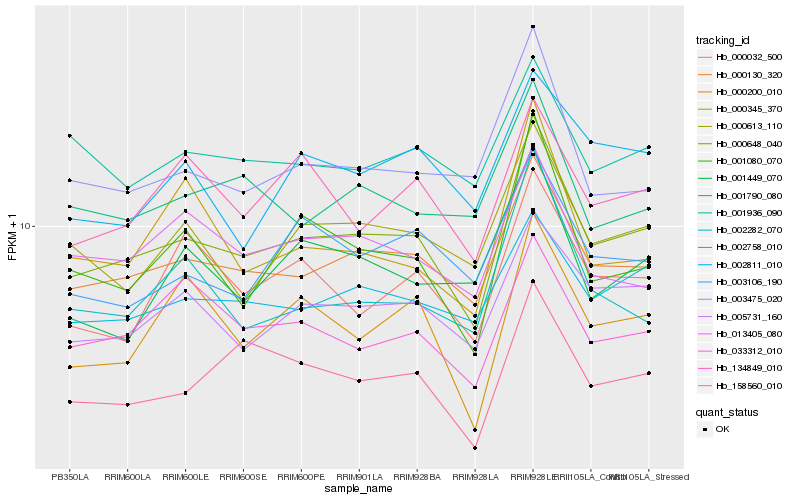
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000648_040 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_000130_320 |
0.0631250469 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 3 |
Hb_005731_160 |
0.0839370293 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_001080_070 |
0.0955140922 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 5 |
Hb_002758_010 |
0.0968314117 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 6 |
Hb_000613_110 |
0.0972459266 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 7 |
Hb_003475_020 |
0.1007384983 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 8 |
Hb_033312_010 |
0.1046795233 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_158560_010 |
0.1056368608 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 10 |
Hb_000032_500 |
0.1067842243 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 11 |
Hb_003106_190 |
0.1075154088 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 12 |
Hb_013405_080 |
0.1076943527 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_002282_070 |
0.108961788 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 14 |
Hb_001790_080 |
0.1095973179 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 15 |
Hb_000200_010 |
0.1099915228 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 16 |
Hb_134849_010 |
0.1102288299 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_001936_090 |
0.1131847522 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000345_370 |
0.1139884592 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 19 |
Hb_002811_010 |
0.1144871139 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 20 |
Hb_001449_070 |
0.1145053069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |