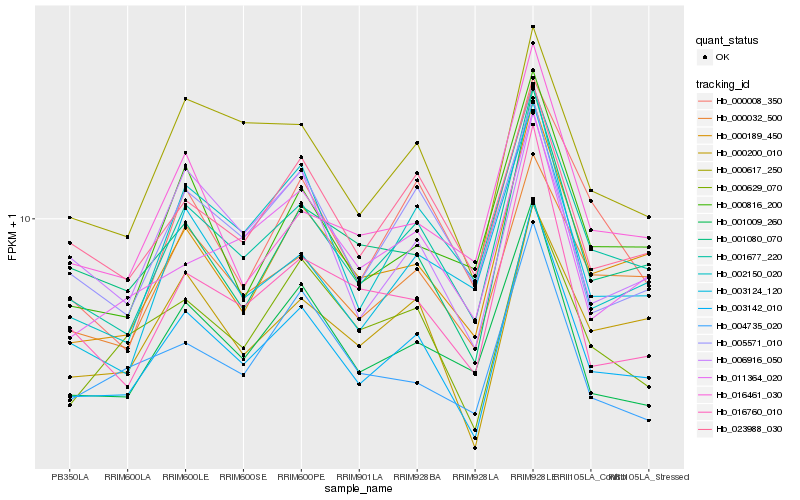
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003142_010 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 2 |
Hb_001677_220 |
0.0556616955 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 3 |
Hb_000189_450 |
0.0822954023 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001080_070 |
0.0880644577 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 5 |
Hb_000200_010 |
0.093147752 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 6 |
Hb_001009_260 |
0.0951950026 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000617_250 |
0.0965829312 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000032_500 |
0.100737998 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 9 |
Hb_016760_010 |
0.1025246143 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_002150_020 |
0.1026558049 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000816_200 |
0.1027826257 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 12 |
Hb_023988_030 |
0.1034828089 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 13 |
Hb_000629_070 |
0.1035302788 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 14 |
Hb_005571_010 |
0.1076345484 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 15 |
Hb_016461_030 |
0.1078682157 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_006916_050 |
0.1088480605 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_004735_020 |
0.1114332969 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 18 |
Hb_003124_120 |
0.111568878 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 19 |
Hb_011364_020 |
0.1118970545 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000008_350 |
0.1132991823 |
- |
- |
- |