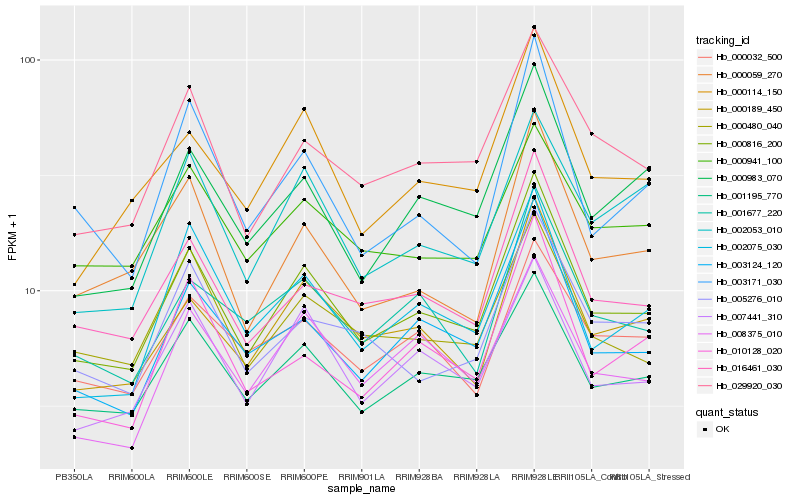
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_200 |
0.0 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 2 |
Hb_016461_030 |
0.067480831 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_001195_770 |
0.0688832009 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 4 |
Hb_003124_120 |
0.069501612 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 5 |
Hb_029920_030 |
0.0723009119 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000189_450 |
0.073528361 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_007441_310 |
0.0772194184 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000114_150 |
0.0790049291 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_000983_070 |
0.0823238564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001677_220 |
0.085105545 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 11 |
Hb_002075_030 |
0.0876825758 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000480_040 |
0.0880298044 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 13 |
Hb_000059_270 |
0.0903424805 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 14 |
Hb_008375_010 |
0.0911543001 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 15 |
Hb_005276_010 |
0.0917738593 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 16 |
Hb_000941_100 |
0.0948418988 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 17 |
Hb_010128_020 |
0.0955187388 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 18 |
Hb_000032_500 |
0.0964663667 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_002053_010 |
0.0978556725 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_003171_030 |
0.0993115043 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |