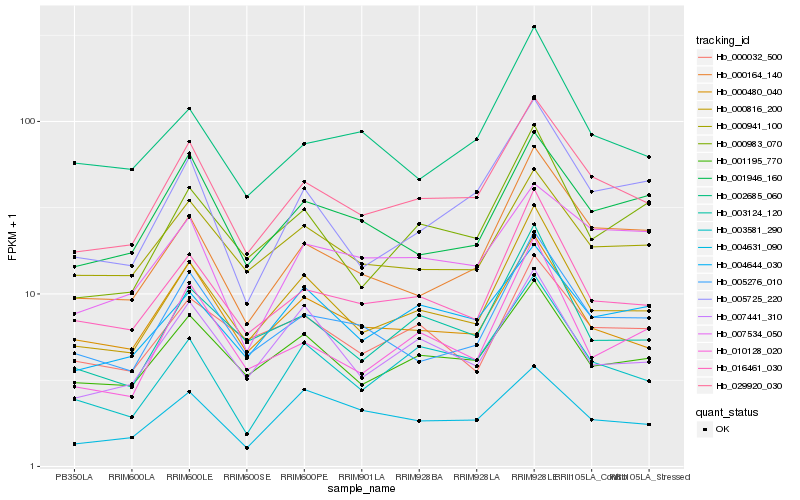
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029920_030 |
0.0 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000816_200 |
0.0723009119 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_016461_030 |
0.084758236 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001195_770 |
0.0859494936 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_005276_010 |
0.0868841388 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 6 |
Hb_000480_040 |
0.0912194138 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 7 |
Hb_000164_140 |
0.0957343945 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_000941_100 |
0.0966894829 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 9 |
Hb_001946_160 |
0.0994650354 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 10 |
Hb_004644_030 |
0.1010506173 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 11 |
Hb_005725_220 |
0.1013008474 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 12 |
Hb_003124_120 |
0.1018385372 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 13 |
Hb_003581_290 |
0.1038110587 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_007534_050 |
0.1074901715 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000983_070 |
0.1077582233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007441_310 |
0.1119509907 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002685_060 |
0.112004214 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 18 |
Hb_010128_020 |
0.11201199 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 19 |
Hb_004631_090 |
0.1120147862 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 20 |
Hb_000032_500 |
0.1127053525 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |