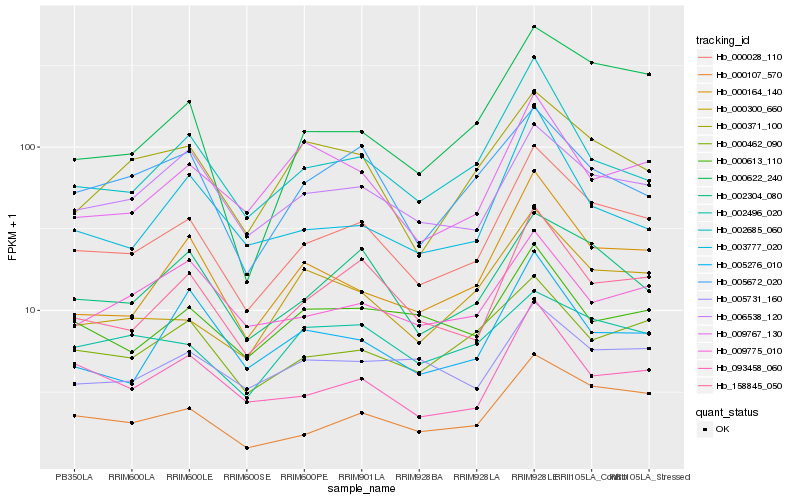
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 2 |
Hb_000107_570 |
0.0741023848 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 3 |
Hb_158845_050 |
0.0842542324 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 4 |
Hb_002304_080 |
0.0939216861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 5 |
Hb_000164_140 |
0.1055194124 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_006538_120 |
0.1083545795 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 7 |
Hb_093458_060 |
0.1100173171 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 8 |
Hb_002685_060 |
0.1113221247 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 9 |
Hb_000462_090 |
0.112901896 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000300_660 |
0.11617631 |
- |
- |
unknown [Glycine max] |
| 11 |
Hb_000622_240 |
0.1183366909 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000371_100 |
0.1183516675 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 13 |
Hb_005672_020 |
0.1194400515 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005731_160 |
0.1230763997 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_000613_110 |
0.1271608871 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 16 |
Hb_009767_130 |
0.128408274 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 17 |
Hb_005276_010 |
0.12901056 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 18 |
Hb_003777_020 |
0.1305223259 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 19 |
Hb_002496_020 |
0.1326967844 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_009775_010 |
0.1328226718 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |