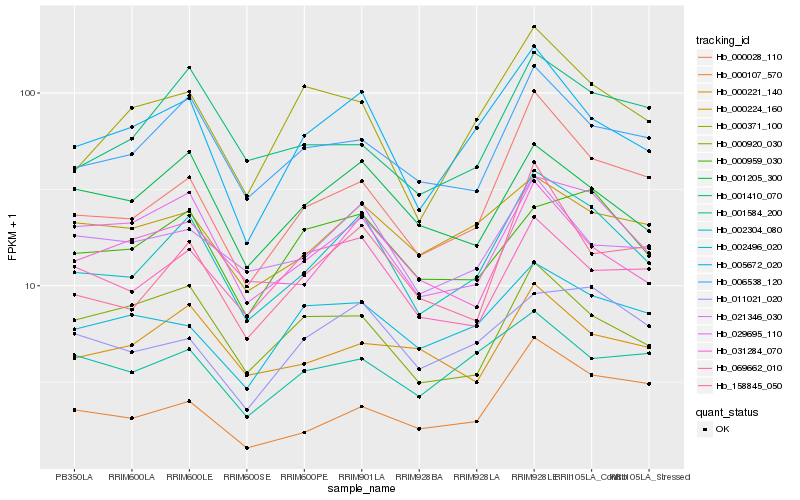
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 2 |
Hb_000028_110 |
0.0939216861 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 3 |
Hb_005672_020 |
0.0950313513 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 4 |
Hb_029695_110 |
0.1018170651 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 5 |
Hb_006538_120 |
0.1032282046 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 6 |
Hb_158845_050 |
0.116099532 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 7 |
Hb_021346_030 |
0.1231586033 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 8 |
Hb_001205_300 |
0.1235523544 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 9 |
Hb_000959_030 |
0.1237410805 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 10 |
Hb_031284_070 |
0.1244665267 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000371_100 |
0.1248511063 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 12 |
Hb_011021_020 |
0.1251220666 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000107_570 |
0.1265539244 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000224_160 |
0.1283253337 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_001410_070 |
0.1291740949 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 16 |
Hb_069662_010 |
0.1339718629 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 17 |
Hb_000221_140 |
0.1342842839 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002496_020 |
0.1352069599 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000920_030 |
0.1362075043 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001584_200 |
0.1387241687 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |