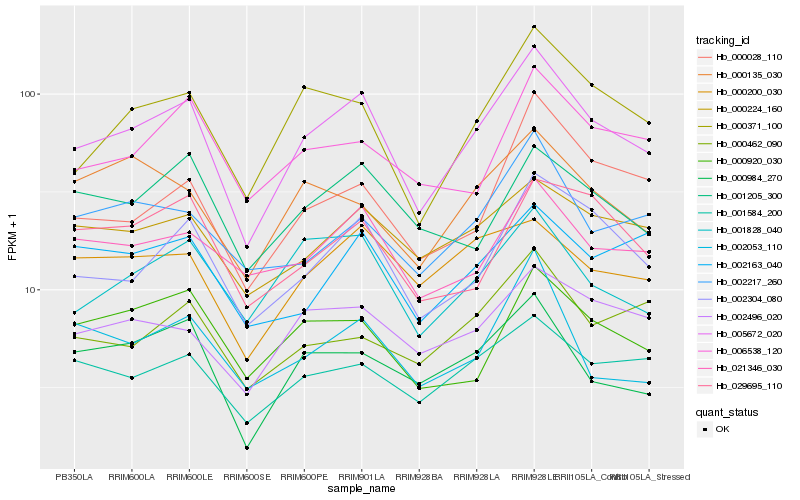
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005672_020 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002304_080 |
0.0950313513 |
- |
- |
PREDICTED: uncharacterized protein LOC105649626 [Jatropha curcas] |
| 3 |
Hb_000371_100 |
0.1004878514 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 4 |
Hb_001828_040 |
0.1095243571 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 5 |
Hb_001584_200 |
0.1110144296 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 6 |
Hb_021346_030 |
0.1148223348 |
- |
- |
PREDICTED: uncharacterized protein LOC105141875 [Populus euphratica] |
| 7 |
Hb_002496_020 |
0.1164314661 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000224_160 |
0.1175929767 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_000984_270 |
0.117975018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000028_110 |
0.1194400515 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 11 |
Hb_000920_030 |
0.1234309613 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_006538_120 |
0.124249804 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000200_030 |
0.1245676111 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001205_300 |
0.1253744344 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 15 |
Hb_002217_260 |
0.1260236237 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 16 |
Hb_029695_110 |
0.1287904687 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 17 |
Hb_000462_090 |
0.1299154379 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002053_110 |
0.1304787989 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000135_030 |
0.1318155468 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002163_040 |
0.1323869163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |