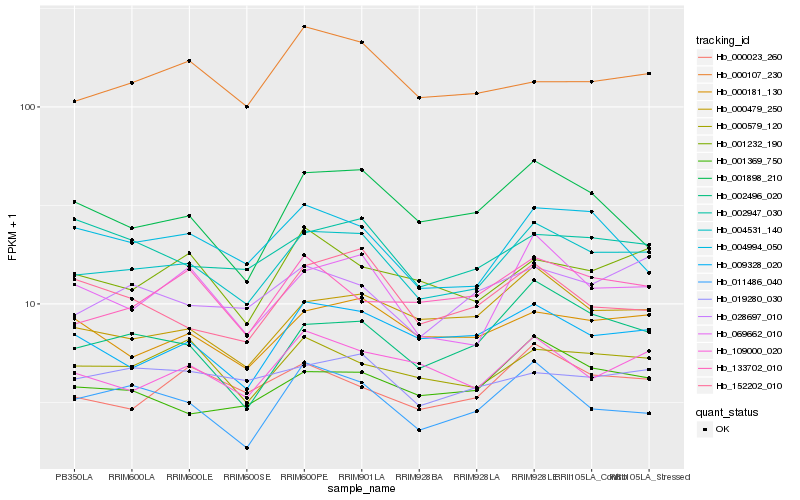
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 2 |
Hb_000479_250 |
0.0805518413 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 3 |
Hb_069662_010 |
0.0820355475 |
- |
- |
PREDICTED: uncharacterized protein LOC103320051 [Prunus mume] |
| 4 |
Hb_000181_130 |
0.0827205674 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_009328_020 |
0.0831778624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_109000_020 |
0.0835164601 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 7 |
Hb_002496_020 |
0.0850452297 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000579_120 |
0.0875756468 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 9 |
Hb_001369_750 |
0.0884887799 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 10 |
Hb_000023_260 |
0.0901014137 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_028697_010 |
0.0908919024 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 12 |
Hb_011486_040 |
0.0923854213 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 13 |
Hb_001232_190 |
0.0937828915 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004994_050 |
0.0944874851 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 15 |
Hb_019280_030 |
0.0959399985 |
- |
- |
- |
| 16 |
Hb_133702_010 |
0.0963900321 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_152202_010 |
0.0967261243 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 18 |
Hb_000107_230 |
0.0983816307 |
- |
- |
unknown [Medicago truncatula] |
| 19 |
Hb_001898_210 |
0.0985412106 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 20 |
Hb_002947_030 |
0.0988656831 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |