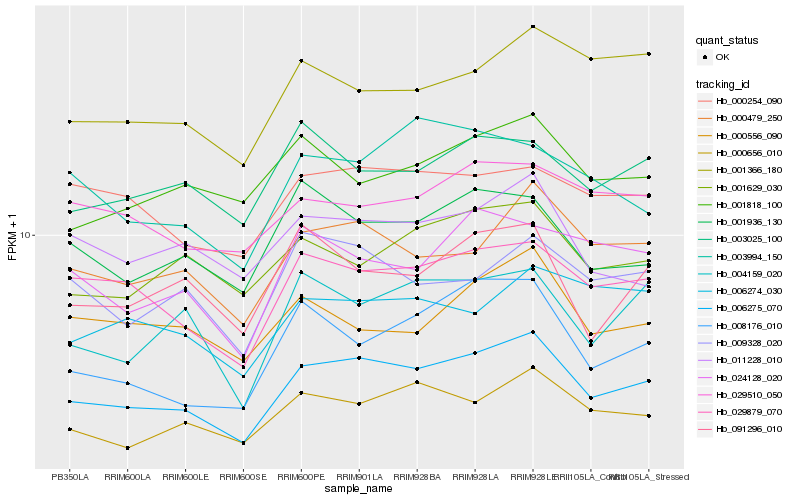
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006275_070 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 2 |
Hb_029879_070 |
0.0700468322 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 3 |
Hb_000479_250 |
0.0722767876 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 4 |
Hb_000656_010 |
0.0771508401 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001936_130 |
0.0864358254 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 6 |
Hb_008176_010 |
0.0870106475 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 7 |
Hb_004159_020 |
0.0871202775 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 8 |
Hb_001629_030 |
0.0881700414 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 9 |
Hb_009328_020 |
0.0888218708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000254_090 |
0.0890376712 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 11 |
Hb_024128_020 |
0.0890450228 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_091296_010 |
0.0908202578 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_001366_180 |
0.091769457 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_011228_010 |
0.0919505661 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003025_100 |
0.0920709763 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_029510_050 |
0.0930427945 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 17 |
Hb_001818_100 |
0.0943367447 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 18 |
Hb_006274_030 |
0.0951811547 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 19 |
Hb_003994_150 |
0.0954554679 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 20 |
Hb_000556_090 |
0.0962302146 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |