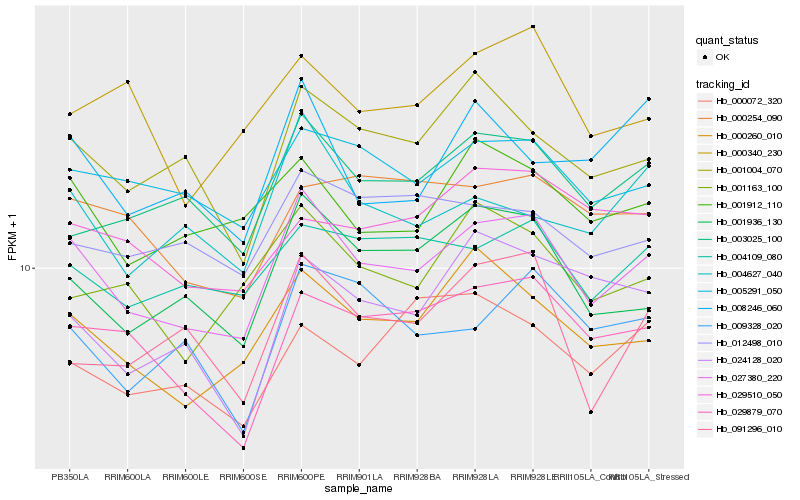
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_220 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 2 |
Hb_000260_010 |
0.0650481358 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 3 |
Hb_001936_130 |
0.0685877359 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 4 |
Hb_005291_050 |
0.0717649513 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_029879_070 |
0.0748123624 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 6 |
Hb_001004_070 |
0.0751145504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001912_110 |
0.0755283602 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_004109_080 |
0.0769012129 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 9 |
Hb_003025_100 |
0.078687469 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004627_040 |
0.0814704806 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 11 |
Hb_001163_100 |
0.0816252257 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_009328_020 |
0.0849457328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012498_010 |
0.0853220393 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 14 |
Hb_000254_090 |
0.085882301 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 15 |
Hb_000340_230 |
0.0886022664 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 16 |
Hb_091296_010 |
0.0895241921 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 17 |
Hb_029510_050 |
0.0896070957 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 18 |
Hb_008246_060 |
0.0904559945 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 19 |
Hb_024128_020 |
0.0921774094 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000072_320 |
0.0927264427 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |