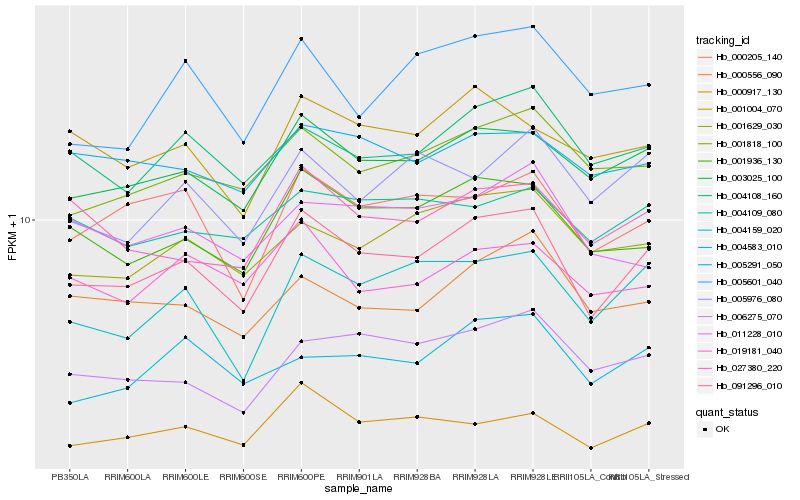
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091296_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_003025_100 |
0.0657188925 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001936_130 |
0.0722227859 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 4 |
Hb_004159_020 |
0.0819119961 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 5 |
Hb_004108_160 |
0.084073523 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 6 |
Hb_001818_100 |
0.0842759306 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 7 |
Hb_000205_140 |
0.0848367565 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 8 |
Hb_001629_030 |
0.0871766113 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 9 |
Hb_027380_220 |
0.0895241921 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 10 |
Hb_006275_070 |
0.0908202578 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 11 |
Hb_000556_090 |
0.0914941913 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001004_070 |
0.0924807443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000917_130 |
0.092944371 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 14 |
Hb_004109_080 |
0.0957580675 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 15 |
Hb_019181_040 |
0.0962740559 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 16 |
Hb_005601_040 |
0.0968381311 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 17 |
Hb_004583_010 |
0.0969711471 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005291_050 |
0.0976279015 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005976_080 |
0.0976893909 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 20 |
Hb_011228_010 |
0.1002964227 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |