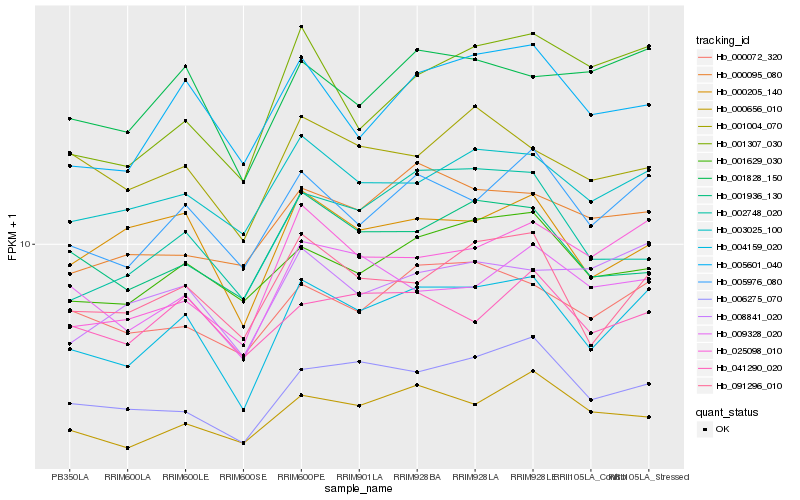
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004159_020 |
0.0 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 2 |
Hb_005976_080 |
0.0703304414 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 3 |
Hb_091296_010 |
0.0819119961 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_006275_070 |
0.0871202775 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 5 |
Hb_003025_100 |
0.0881764943 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000072_320 |
0.0894205454 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 7 |
Hb_001828_150 |
0.0910163696 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005601_040 |
0.0916874692 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_025098_010 |
0.0934879152 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_001629_030 |
0.0951230108 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 11 |
Hb_000656_010 |
0.0958943965 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000205_140 |
0.0976346272 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 13 |
Hb_008841_020 |
0.1000773152 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009328_020 |
0.1030161309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000095_080 |
0.1036195487 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 16 |
Hb_001936_130 |
0.1038486581 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 17 |
Hb_002748_020 |
0.104023315 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 18 |
Hb_001307_030 |
0.1055803266 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001004_070 |
0.1058440134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_041290_020 |
0.1062270714 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |