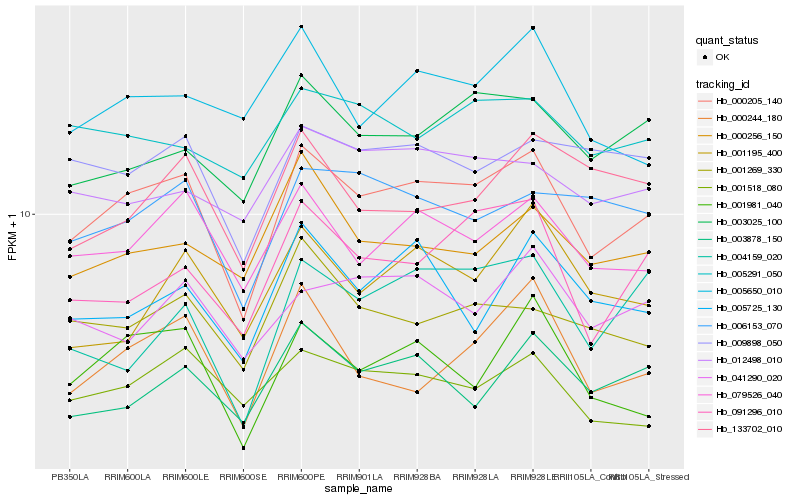
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 2 |
Hb_079526_040 |
0.0744585003 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 3 |
Hb_001981_040 |
0.081830903 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 4 |
Hb_091296_010 |
0.0848367565 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_003025_100 |
0.0864413626 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006153_070 |
0.0866689183 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001518_080 |
0.088040269 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 8 |
Hb_009898_050 |
0.0891048847 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 9 |
Hb_000244_180 |
0.0903438467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003878_150 |
0.0912535578 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 11 |
Hb_041290_020 |
0.0915948994 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_001269_330 |
0.0924510886 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 13 |
Hb_012498_010 |
0.0938567099 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 14 |
Hb_000256_150 |
0.0943175385 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 15 |
Hb_005725_130 |
0.0944501562 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_005291_050 |
0.0955564942 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004159_020 |
0.0976346272 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 18 |
Hb_005650_010 |
0.098544107 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_001195_400 |
0.10021408 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 20 |
Hb_133702_010 |
0.1007815808 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |