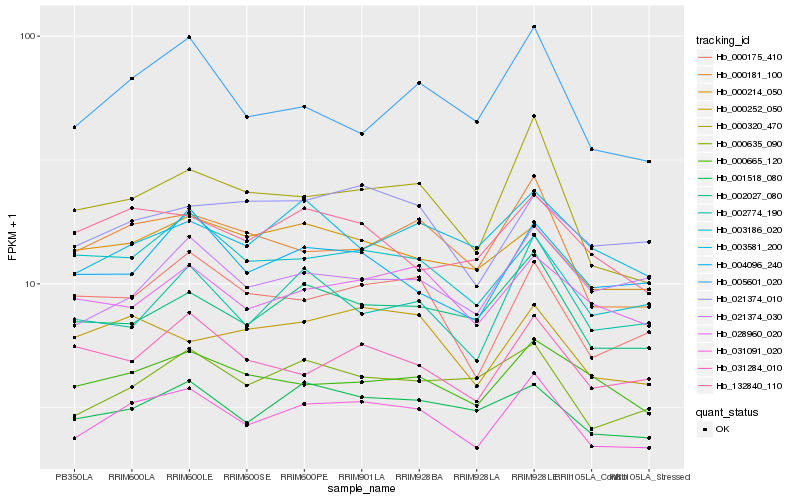
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031091_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_002027_080 |
0.0750157618 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 3 |
Hb_000181_100 |
0.0753577227 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 4 |
Hb_000320_470 |
0.0757106073 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 5 |
Hb_001518_080 |
0.077128791 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 6 |
Hb_003186_020 |
0.0771370753 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000635_090 |
0.0802649488 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 8 |
Hb_000175_410 |
0.0813611881 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_028960_020 |
0.0816334144 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000665_120 |
0.0849926335 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 11 |
Hb_000214_050 |
0.0851504427 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000252_050 |
0.0861616999 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 13 |
Hb_004096_240 |
0.0861781402 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 14 |
Hb_005601_020 |
0.0883170596 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_021374_010 |
0.0884224863 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 16 |
Hb_002774_190 |
0.0892444686 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 17 |
Hb_132840_110 |
0.0896124751 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_031284_010 |
0.0906827498 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 19 |
Hb_021374_030 |
0.0907804387 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 20 |
Hb_003581_200 |
0.091300806 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |