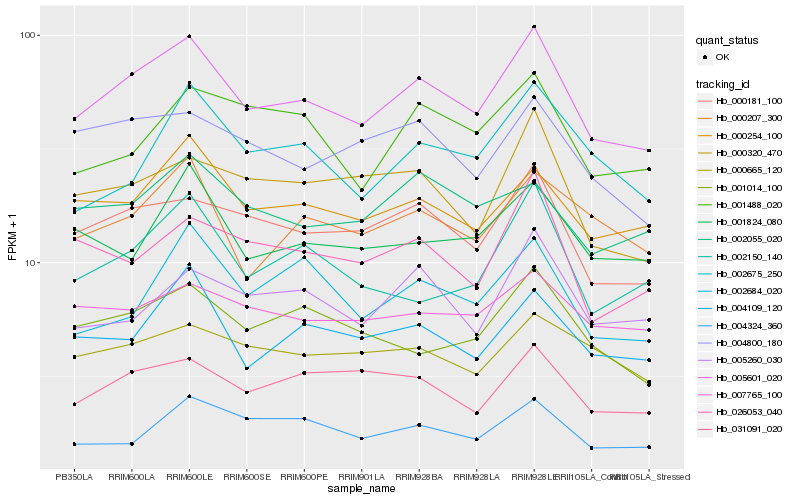
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005601_020 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_000181_100 |
0.0714906111 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 3 |
Hb_000254_100 |
0.083195445 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 4 |
Hb_031091_020 |
0.0883170596 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 5 |
Hb_004109_120 |
0.0884484641 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001014_100 |
0.090860756 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 7 |
Hb_000207_300 |
0.0922454534 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000320_470 |
0.0944364468 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 9 |
Hb_004800_180 |
0.0954948514 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 10 |
Hb_004324_360 |
0.0957565633 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005260_030 |
0.0965690765 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 12 |
Hb_001824_080 |
0.0970990804 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 13 |
Hb_007765_100 |
0.0971803943 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 14 |
Hb_002150_140 |
0.0977572461 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 15 |
Hb_000665_120 |
0.0978520639 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 16 |
Hb_026053_040 |
0.0983158633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002684_020 |
0.0990089077 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 18 |
Hb_002055_020 |
0.0990886416 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 19 |
Hb_002675_250 |
0.0994422133 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_001488_020 |
0.0999606895 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |