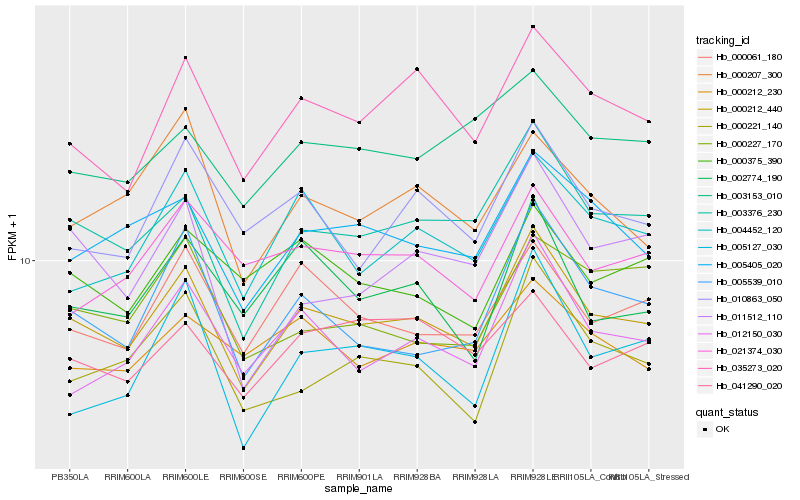
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_440 |
0.0 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_003376_230 |
0.0566183134 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 3 |
Hb_035273_020 |
0.0660885843 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005405_020 |
0.0670649788 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_000061_180 |
0.0719648967 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 6 |
Hb_005539_010 |
0.0726139355 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 7 |
Hb_004452_120 |
0.0753823005 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 8 |
Hb_012150_030 |
0.0763986163 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000221_140 |
0.0798990372 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_010863_050 |
0.0806321491 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 11 |
Hb_002774_190 |
0.0815019931 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 12 |
Hb_003153_010 |
0.0818740148 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 13 |
Hb_000207_300 |
0.0833429005 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000212_230 |
0.0834586165 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 15 |
Hb_041290_020 |
0.0839355778 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000375_390 |
0.0851704499 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 17 |
Hb_011512_110 |
0.087517101 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_005127_030 |
0.0885569469 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_021374_030 |
0.0892355156 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 20 |
Hb_000227_170 |
0.0905441448 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |