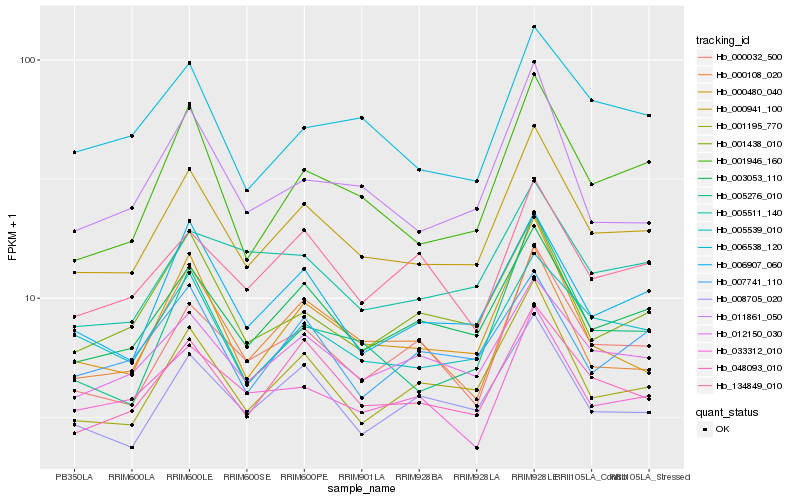
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000941_100 |
0.0 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 2 |
Hb_003053_110 |
0.0516901971 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_048093_010 |
0.0610960874 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001195_770 |
0.0638837933 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_012150_030 |
0.0648816124 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_006907_060 |
0.0671682466 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_005539_010 |
0.0729094722 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 8 |
Hb_008705_020 |
0.0733915083 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_001946_160 |
0.0742655308 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 10 |
Hb_000032_500 |
0.0761408562 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 11 |
Hb_000480_040 |
0.0762723981 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 12 |
Hb_001438_010 |
0.0813446621 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005276_010 |
0.0824924225 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 14 |
Hb_006538_120 |
0.085388957 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 15 |
Hb_033312_010 |
0.0861500136 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_011861_050 |
0.0862424943 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_134849_010 |
0.0867385082 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 18 |
Hb_005511_140 |
0.0874863464 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 19 |
Hb_007741_110 |
0.087781793 |
- |
- |
- |
| 20 |
Hb_000108_020 |
0.0881347313 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |