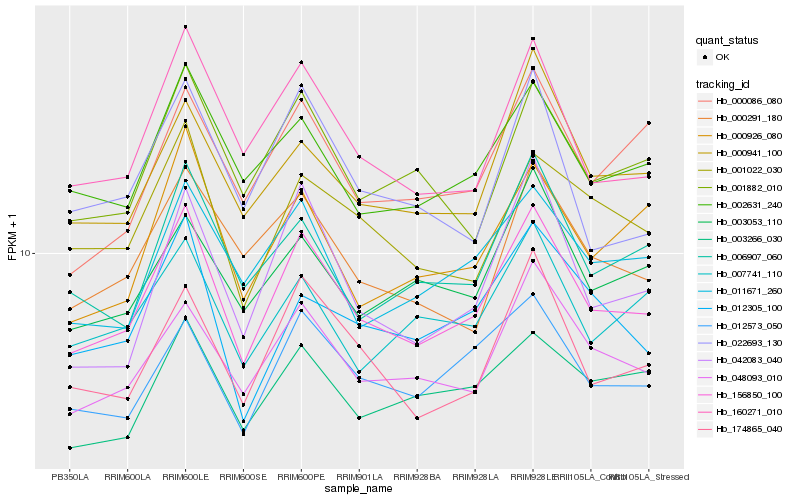
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156850_100 |
0.0 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 2 |
Hb_160271_010 |
0.0717208487 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_048093_010 |
0.0742033305 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_174865_040 |
0.0797215712 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002631_240 |
0.0813007638 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 6 |
Hb_012573_050 |
0.0827425412 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 7 |
Hb_001882_010 |
0.0854169947 |
- |
- |
- |
| 8 |
Hb_011671_260 |
0.0868769751 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000941_100 |
0.0882667391 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 10 |
Hb_001022_030 |
0.08898019 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 11 |
Hb_006907_060 |
0.0893501695 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000291_180 |
0.0898522221 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 13 |
Hb_042083_040 |
0.0903397817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000926_080 |
0.0928105477 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 15 |
Hb_003266_030 |
0.0938941212 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 16 |
Hb_000086_080 |
0.0939645919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003053_110 |
0.094574293 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_022693_130 |
0.0959627725 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_007741_110 |
0.0962488418 |
- |
- |
- |
| 20 |
Hb_012305_100 |
0.0962540894 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |